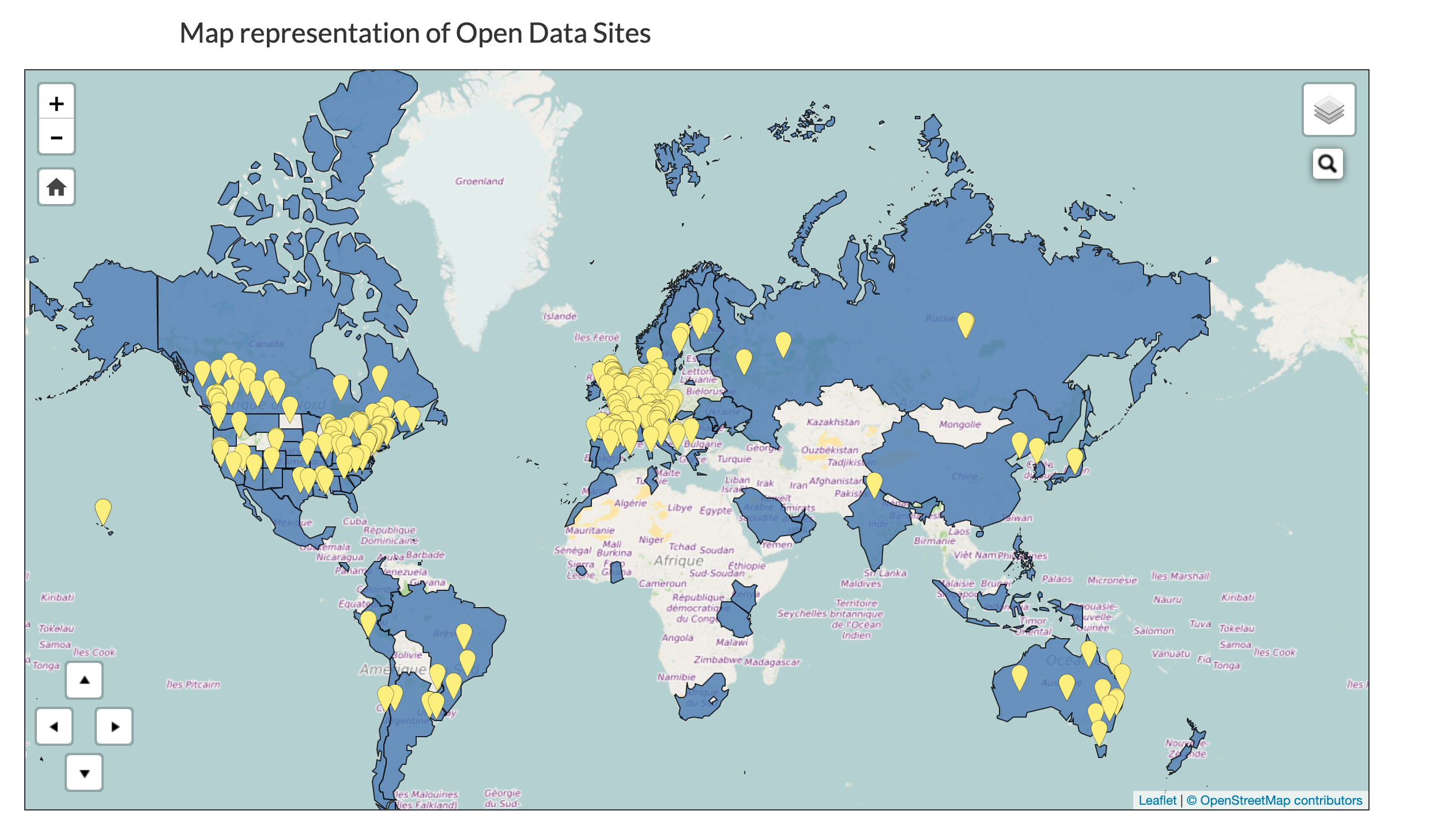
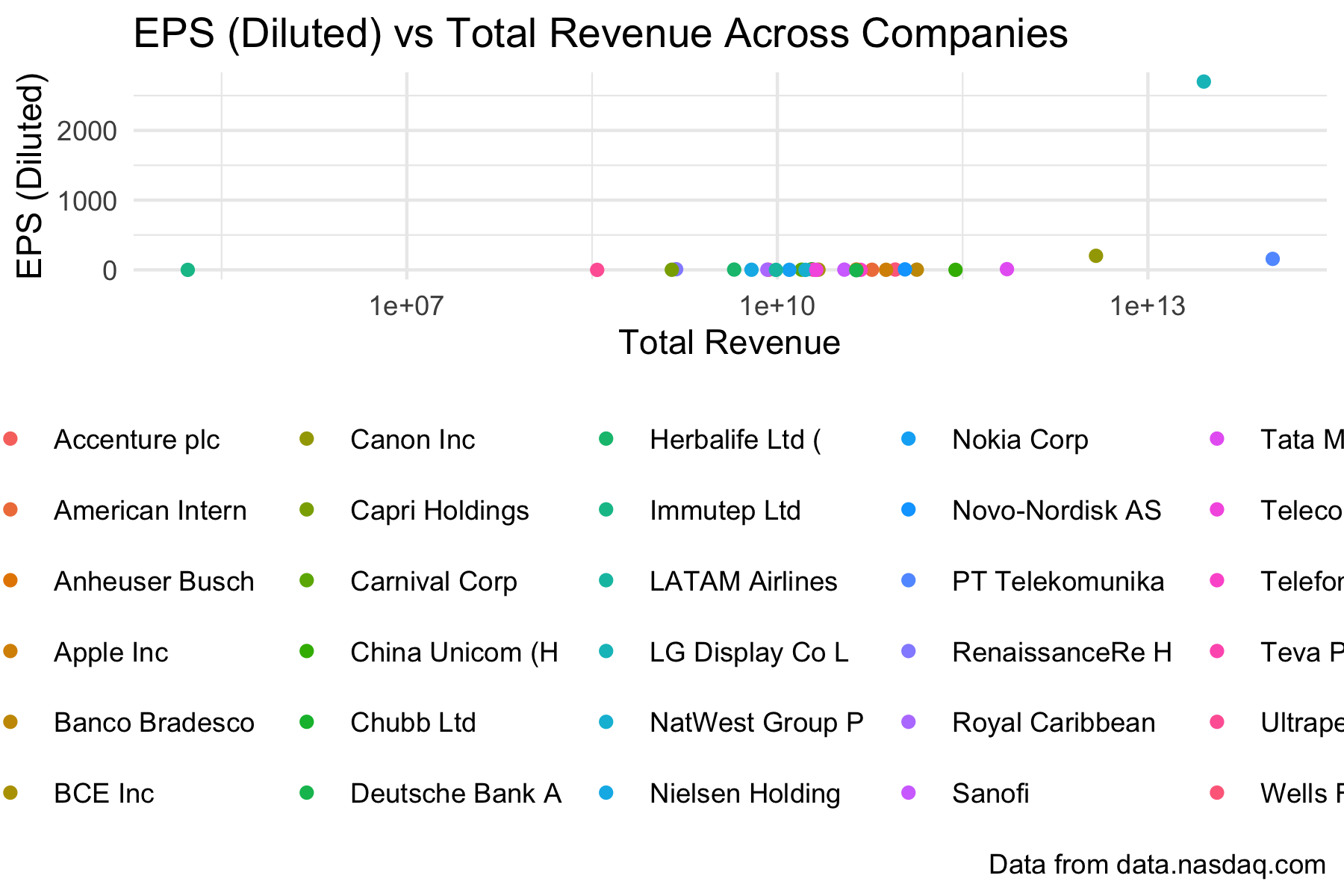
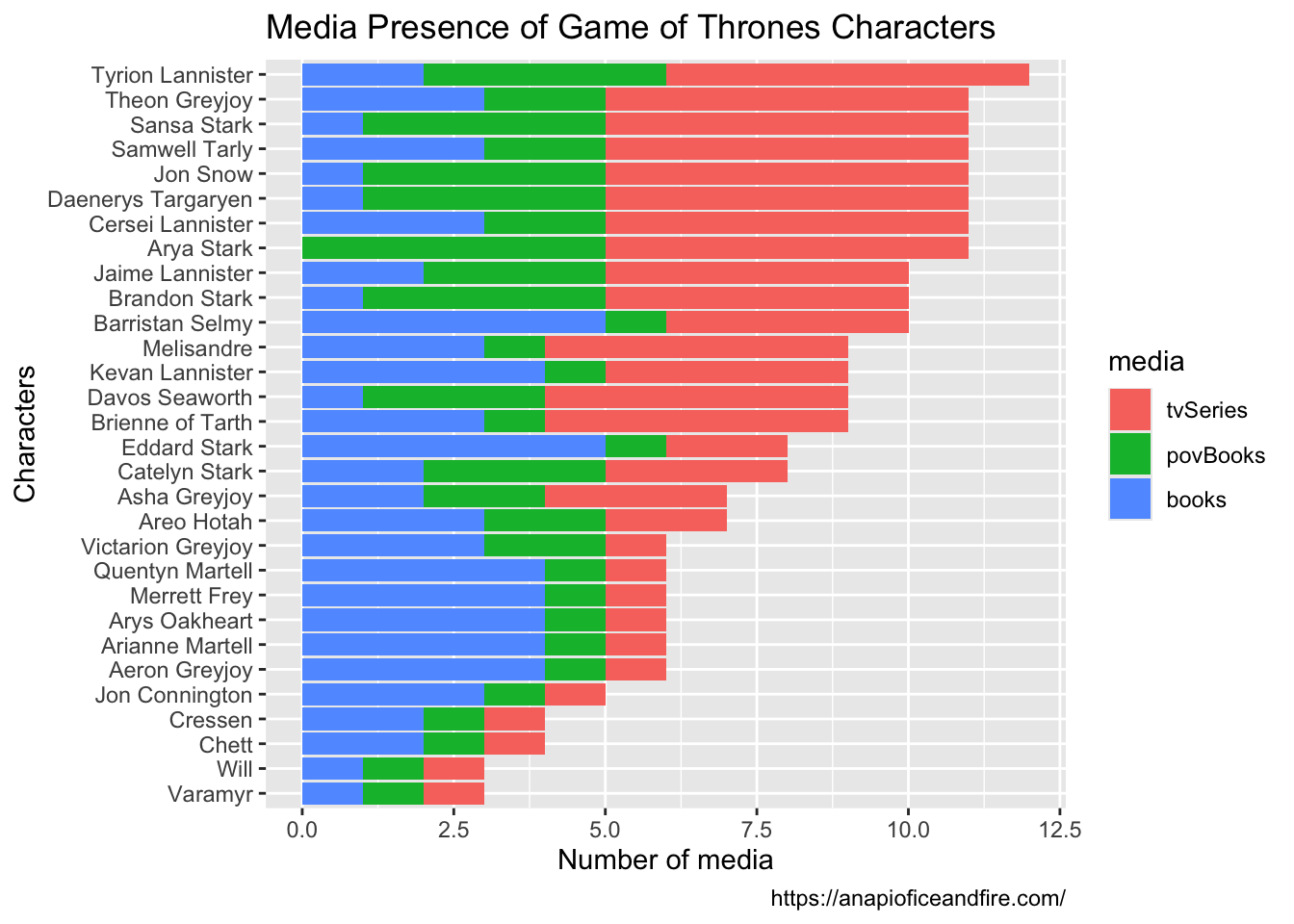
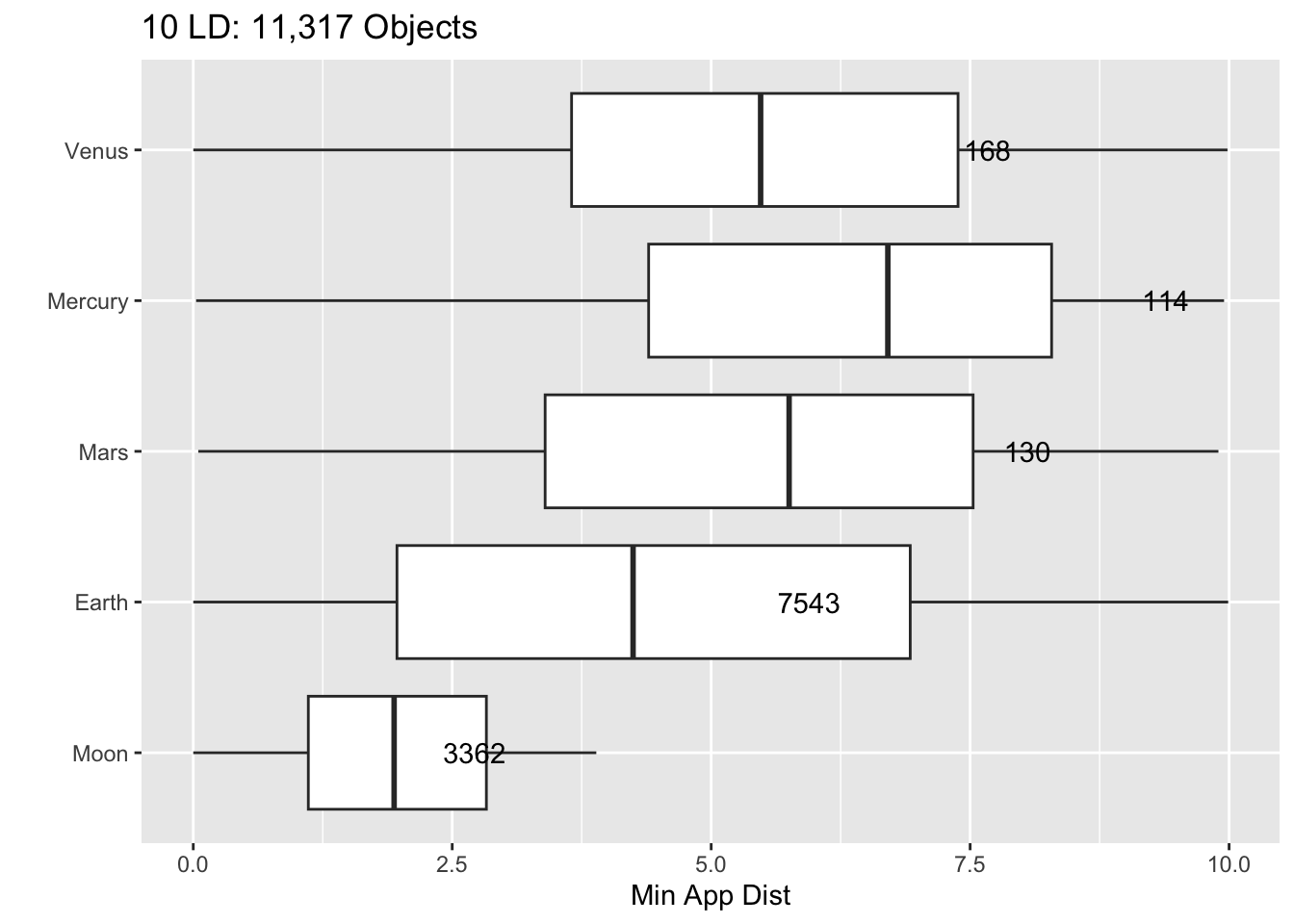
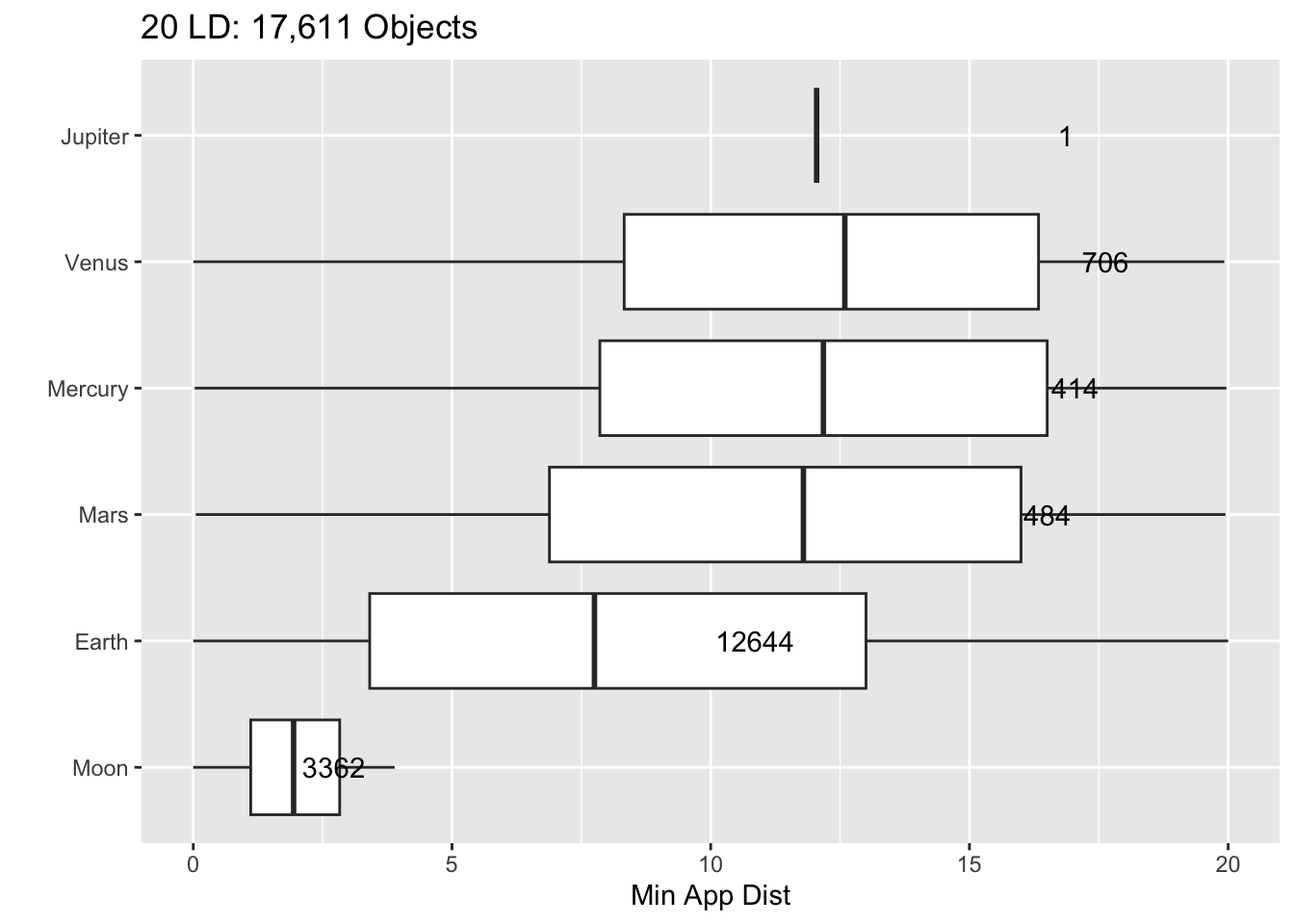
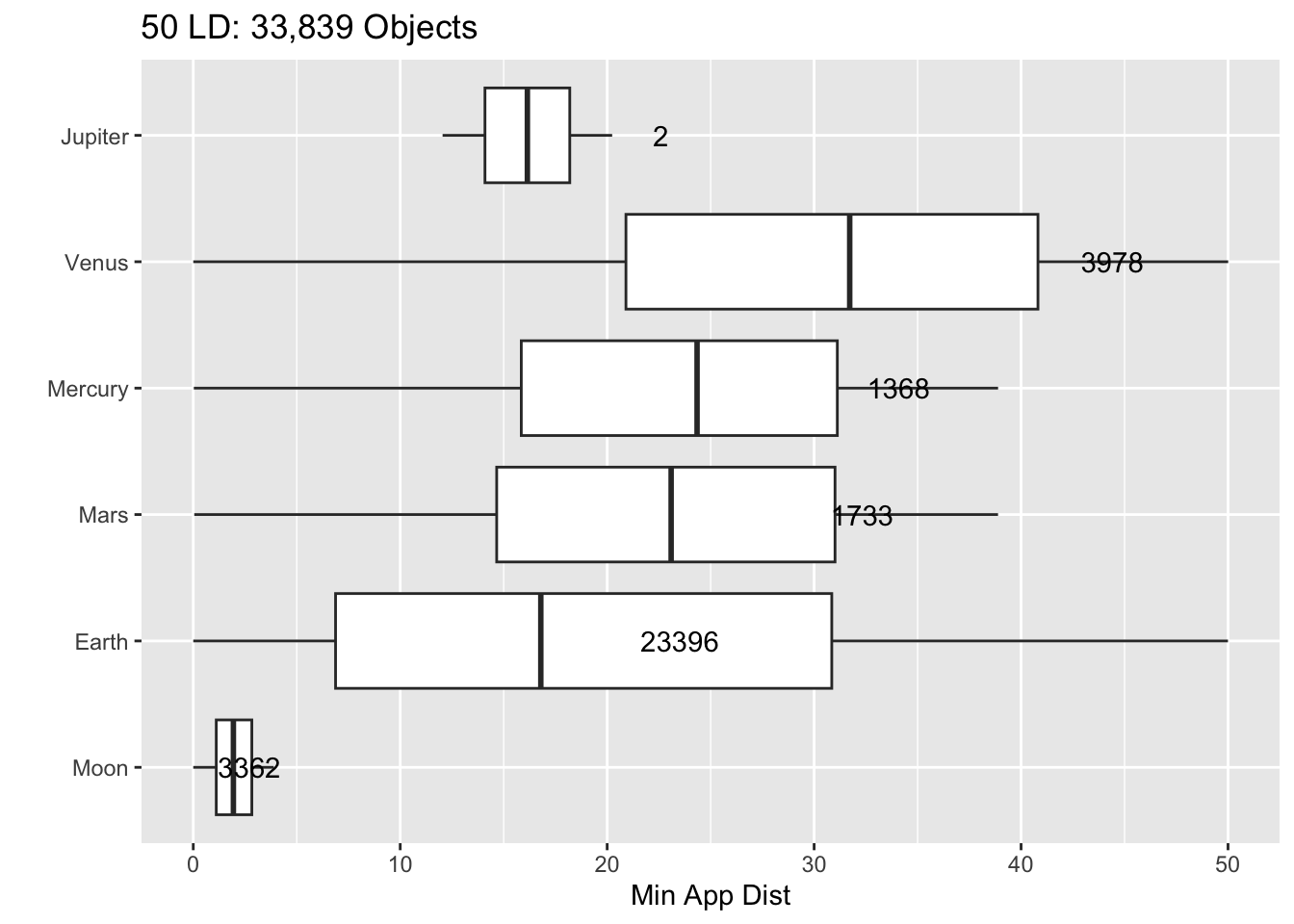
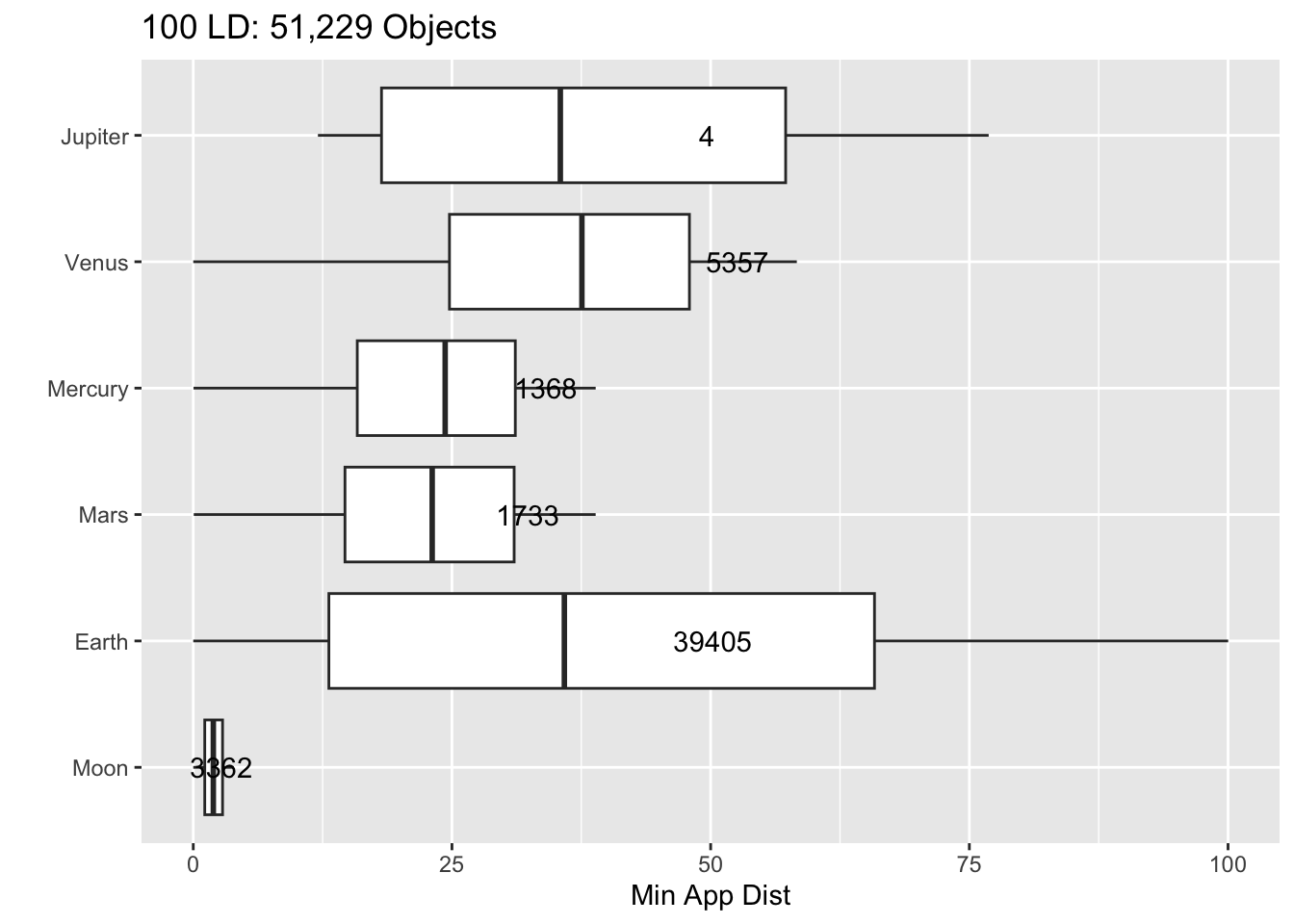
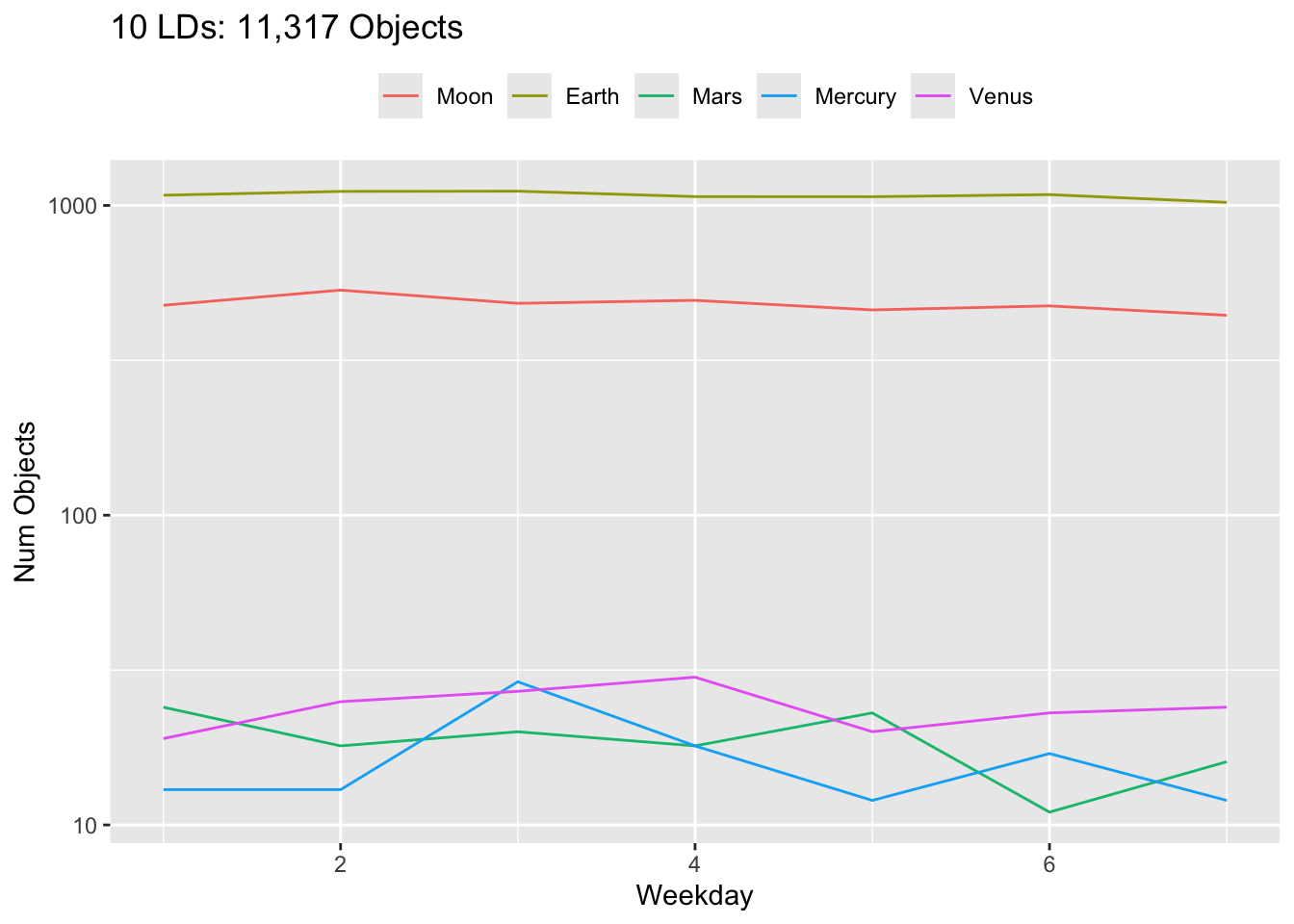
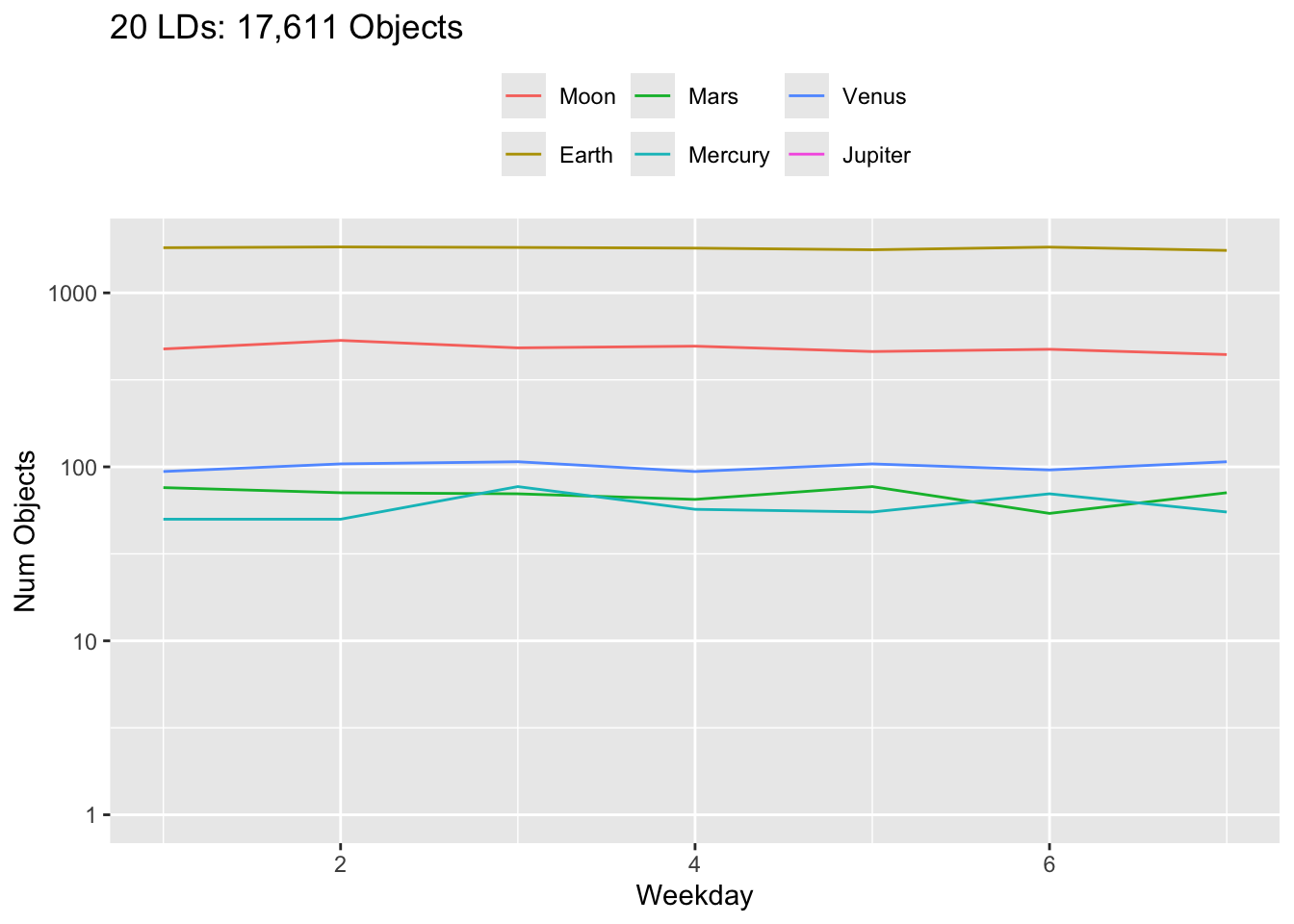
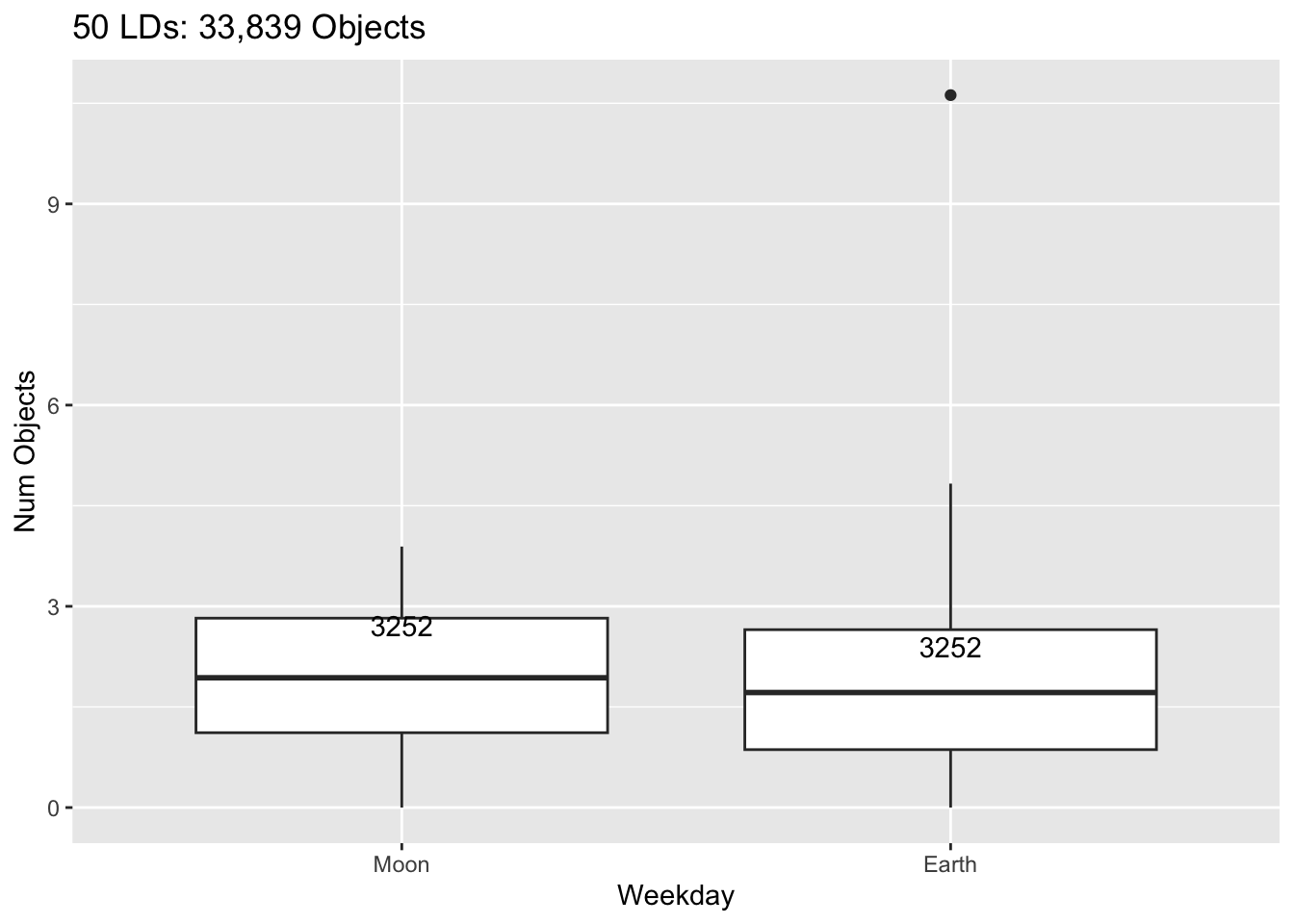
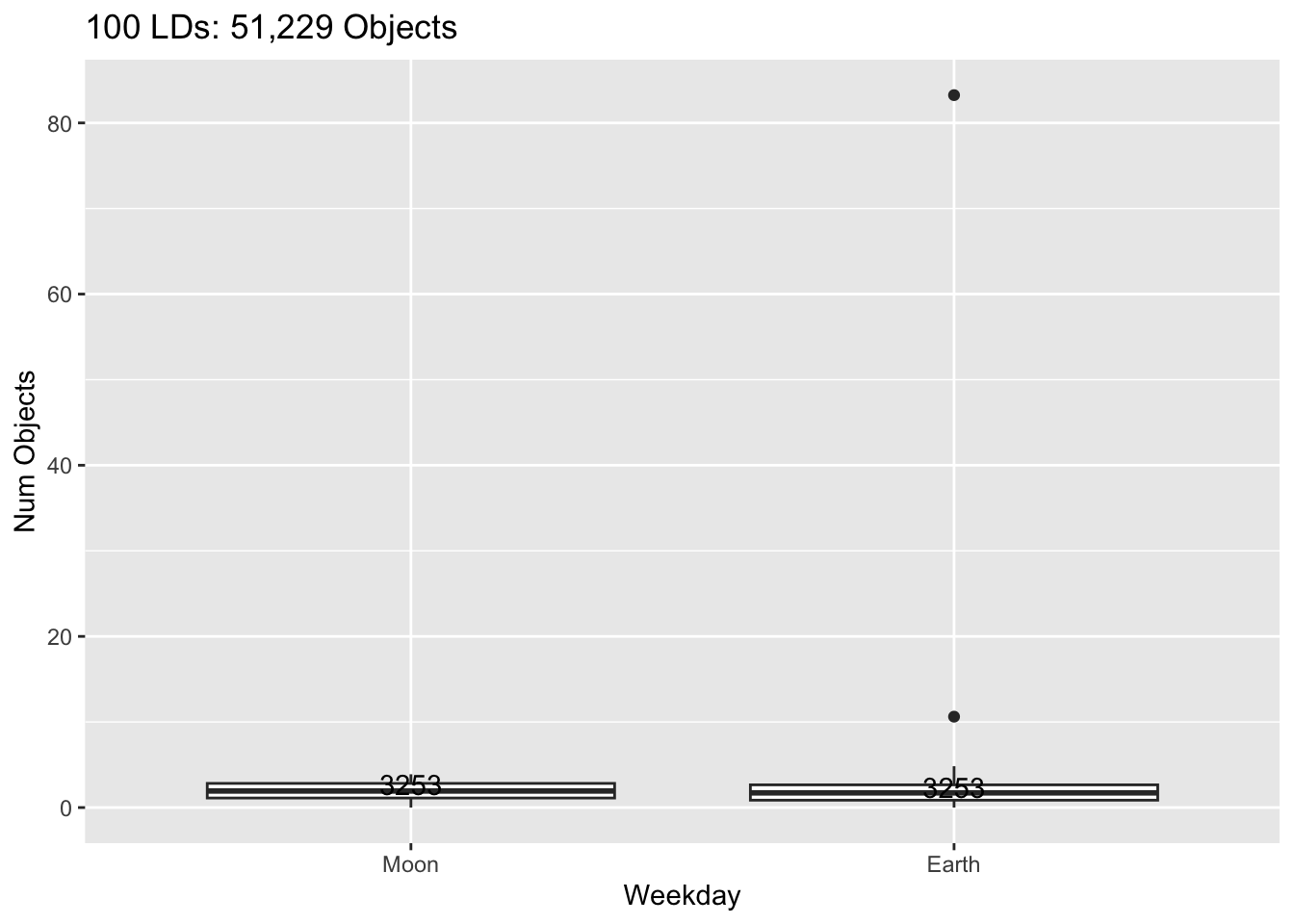
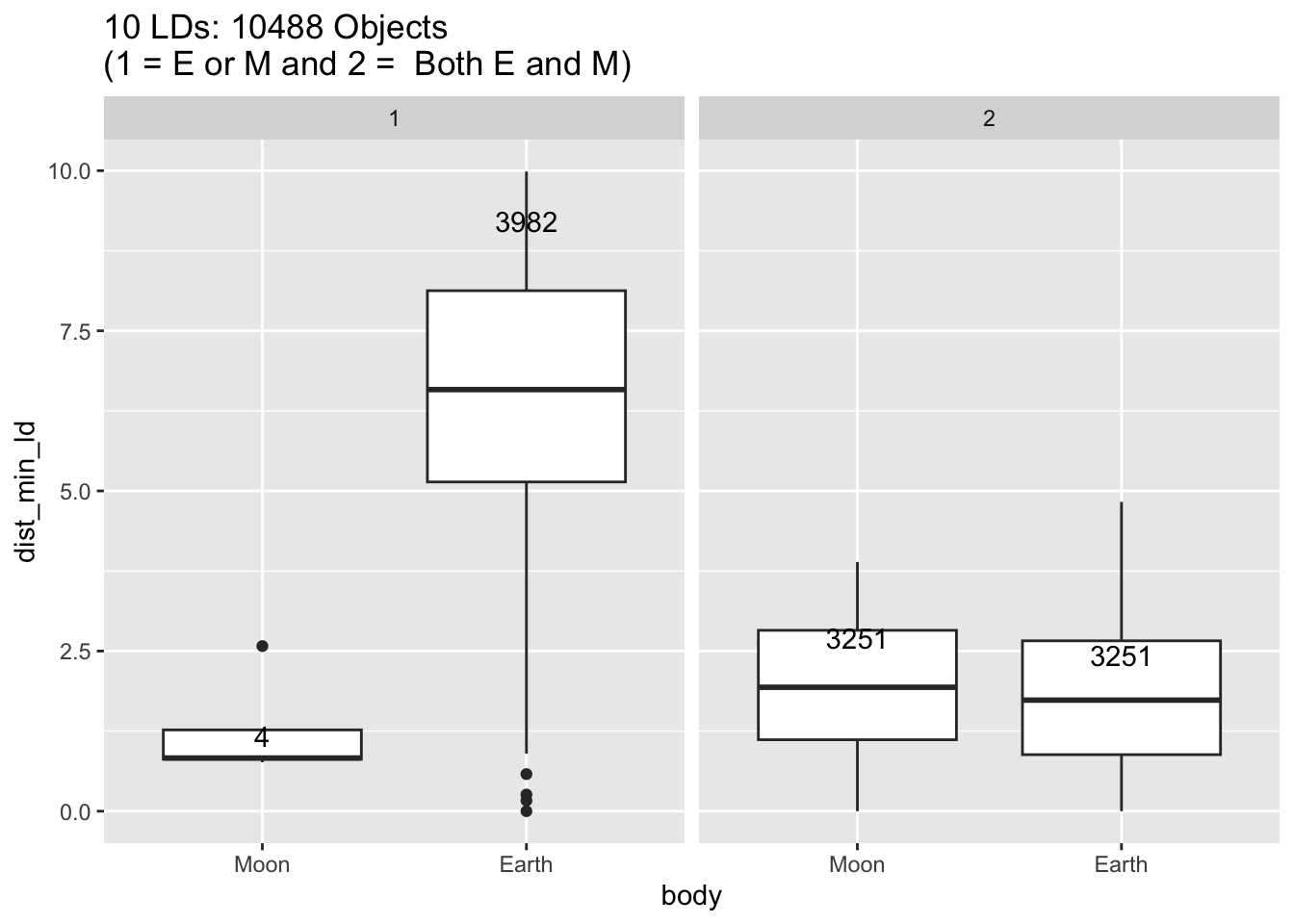
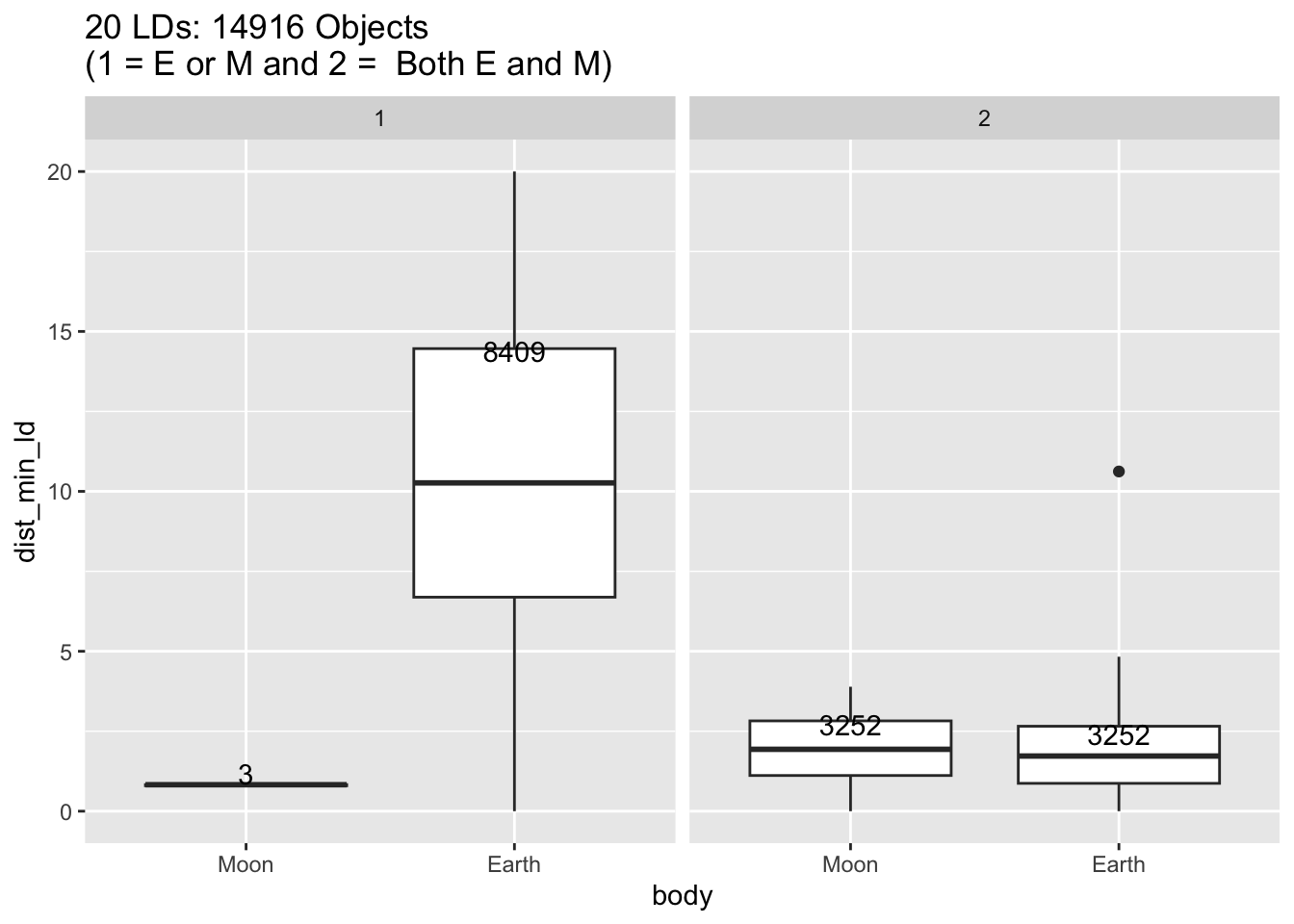
Rows: 30
Columns: 18
$ url <chr> "https://www.anapioficeandfire.com/api/characters/1022", "…
$ id <int> 1022, 1052, 1074, 1109, 1166, 1267, 1295, 130, 1303, 1319,…
$ name <chr> "Theon Greyjoy", "Tyrion Lannister", "Victarion Greyjoy", …
$ gender <chr> "Male", "Male", "Male", "Male", "Male", "Male", "Male", "F…
$ culture <chr> "Ironborn", "", "Ironborn", "", "Norvoshi", "", "", "Dorni…
$ born <chr> "In 278 AC or 279 AC, at Pyke", "In 273 AC, at Casterly Ro…
$ died <chr> "", "", "", "In 297 AC, at Haunted Forest", "", "In 299 AC…
$ alive <lgl> TRUE, TRUE, TRUE, FALSE, TRUE, FALSE, FALSE, TRUE, TRUE, T…
$ titles <list> <"Prince of Winterfell", "Lord of the Iron Islands (by la…
$ aliases <list> <"Prince of Fools", "Theon Turncloak", "Reek", "Theon Kin…
$ father <chr> "", "", "", "", "", "", "", "", "", "", "", "", "", "", ""…
$ mother <chr> "", "", "", "", "", "", "", "", "", "", "", "", "", "", ""…
$ spouse <chr> "", "https://www.anapioficeandfire.com/api/characters/2044…
$ allegiances <list> "House Greyjoy of Pyke", "House Lannister of Casterly Roc…
$ books <list> <"A Game of Thrones", "A Storm of Swords", "A Feast for C…
$ povBooks <list> <"A Clash of Kings", "A Dance with Dragons">, <"A Game of…
$ tvSeries <list> <"Season 1", "Season 2", "Season 3", "Season 4", "Season …
$ playedBy <list> "Alfie Allen", "Peter Dinklage", "", "Bronson Webb", "DeO…