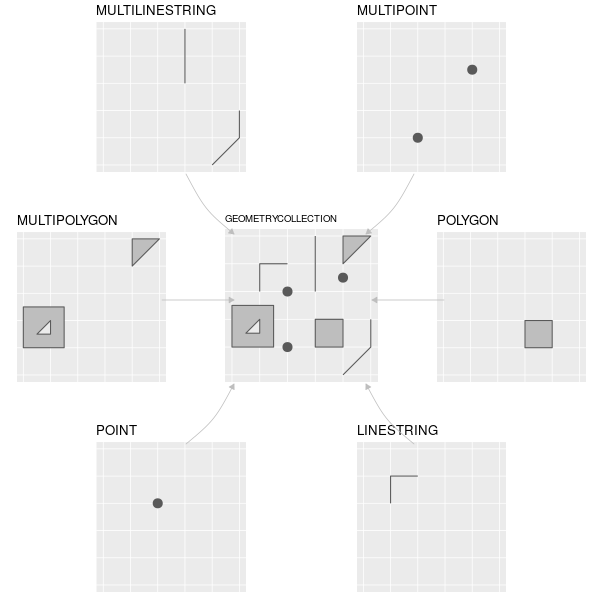
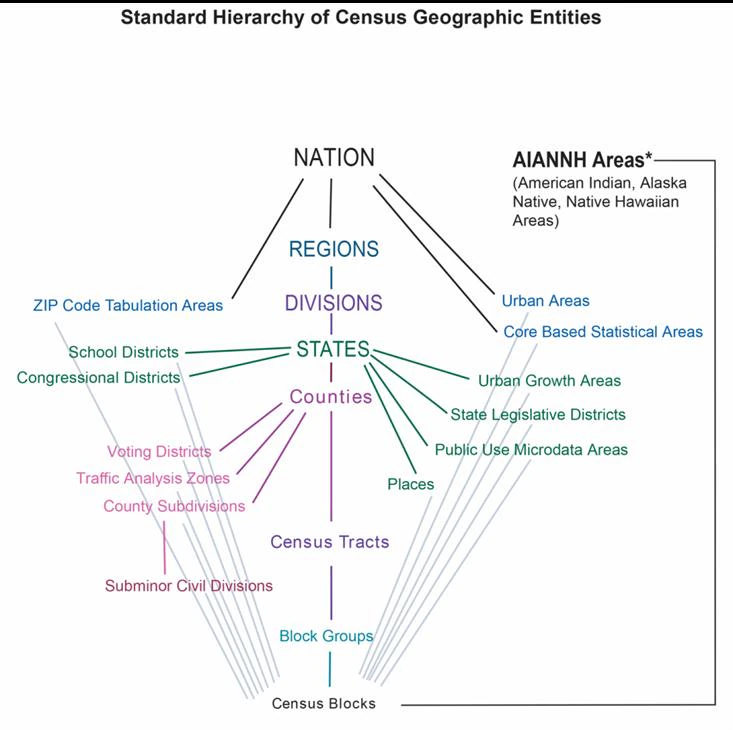
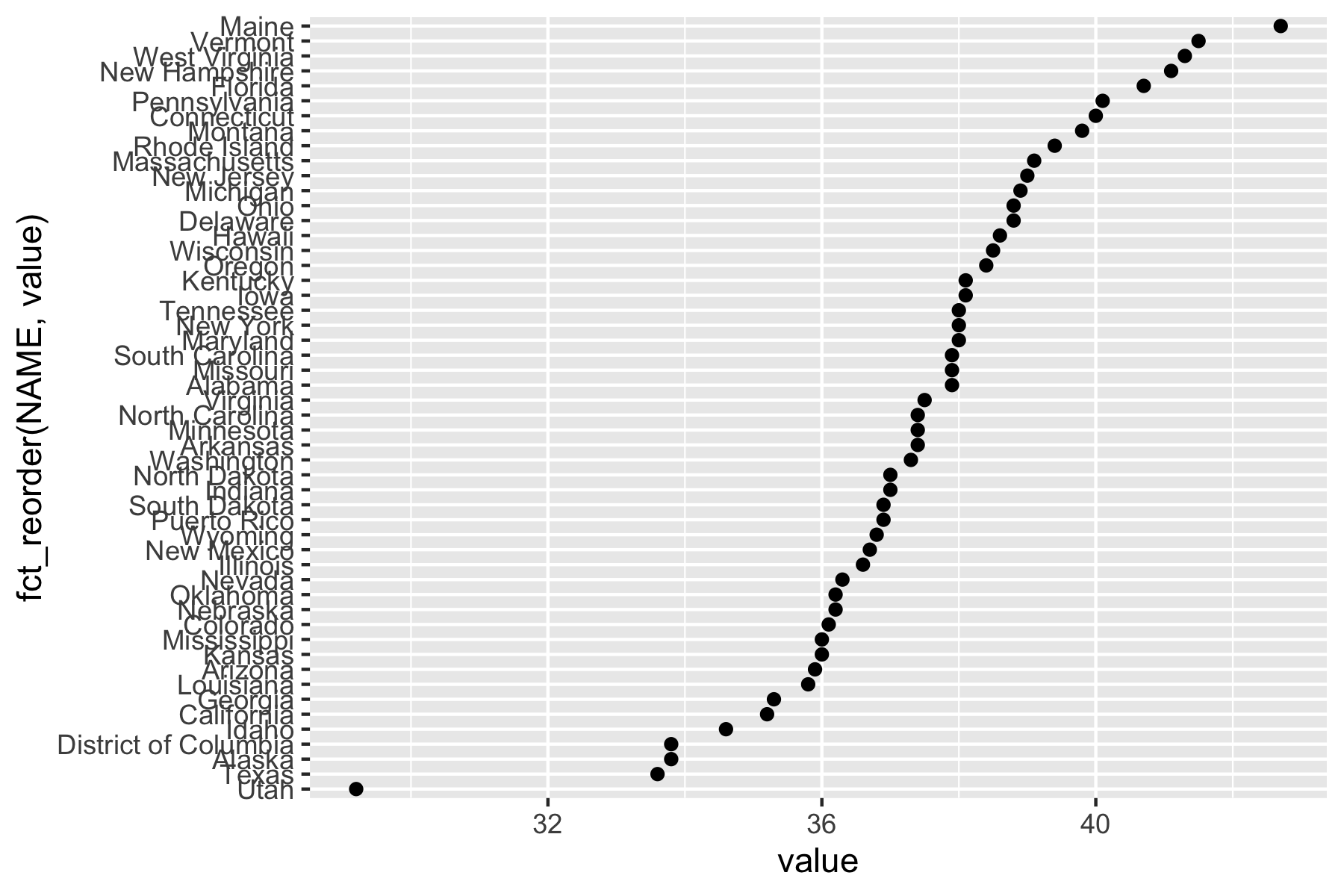
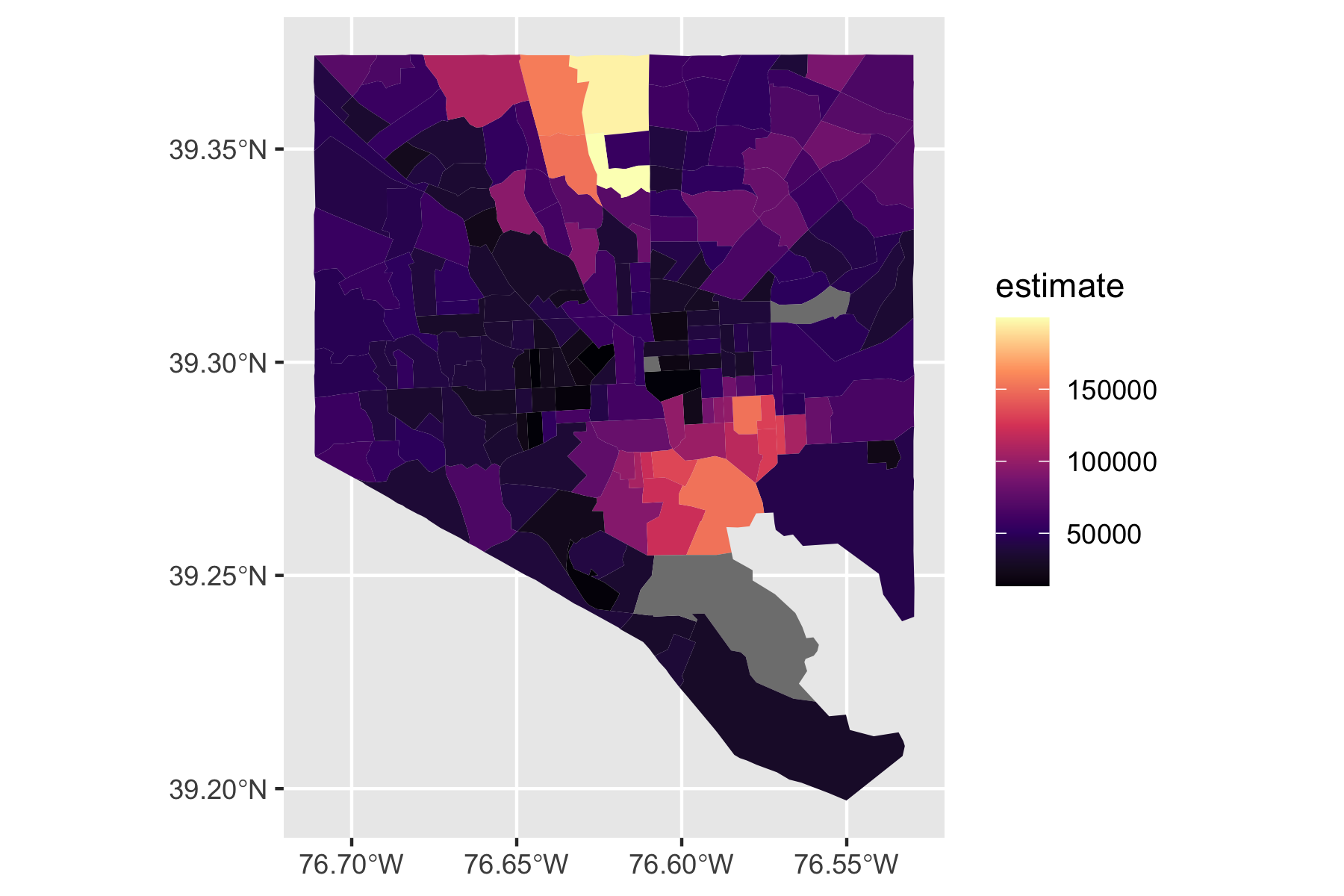
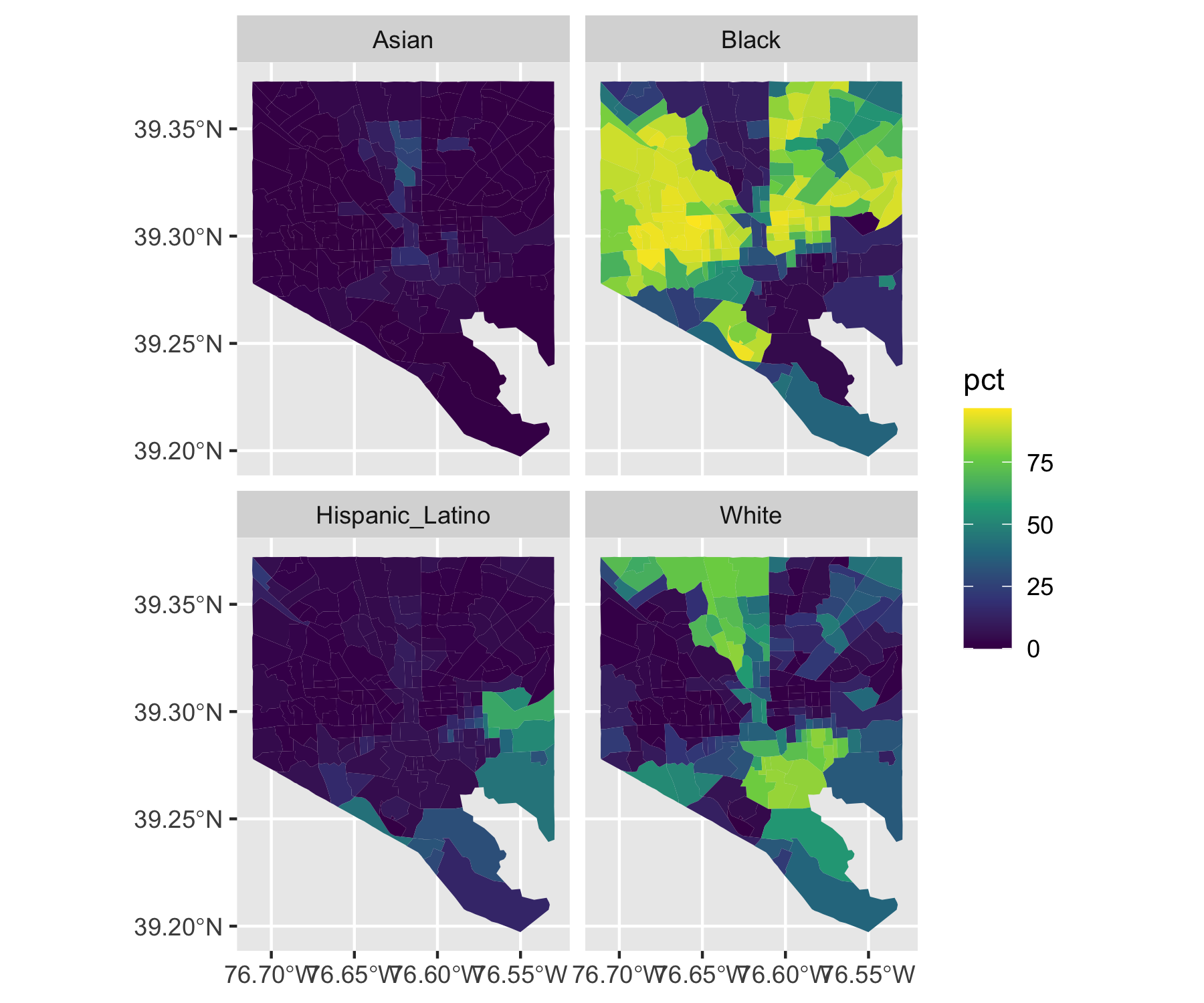
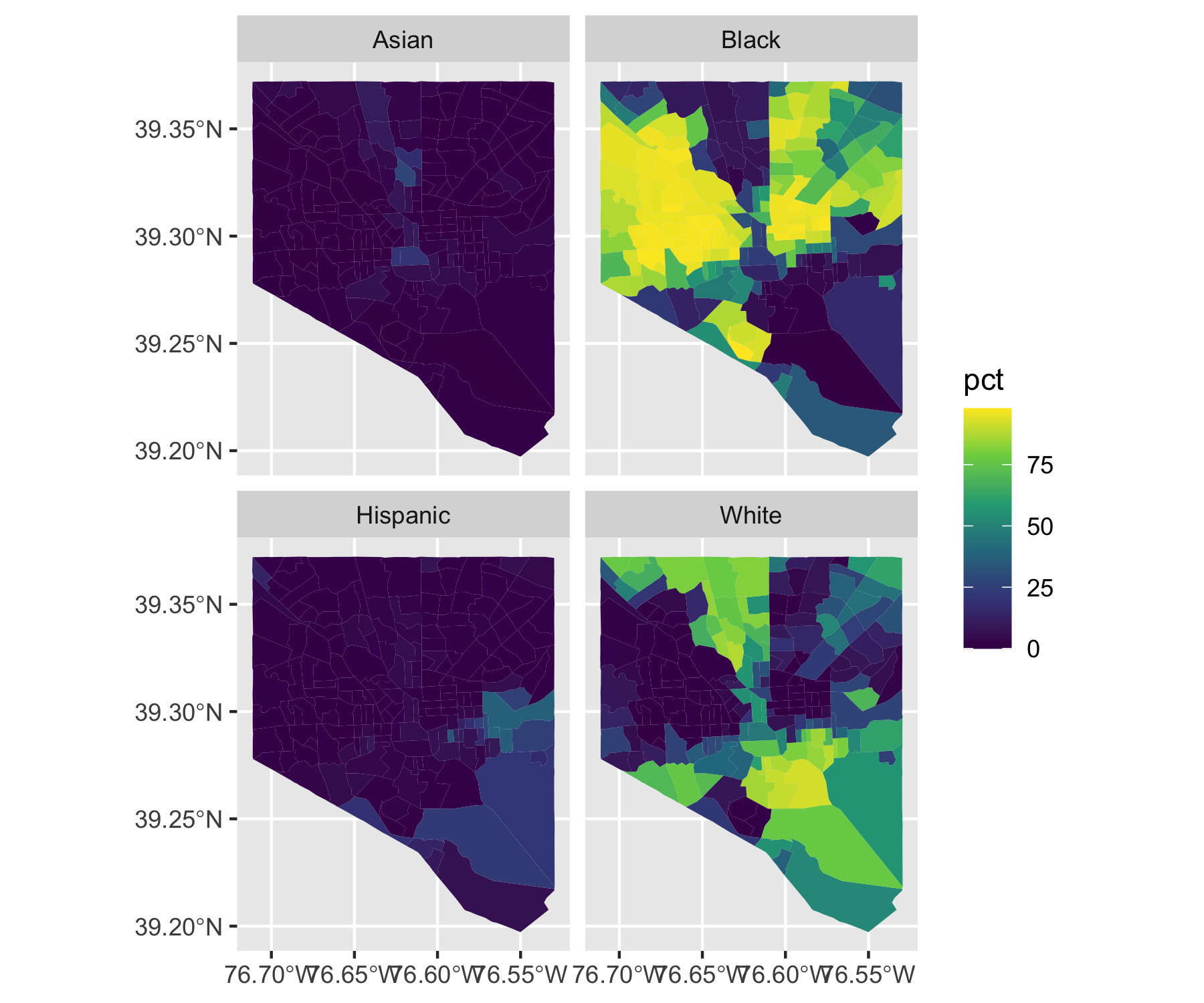
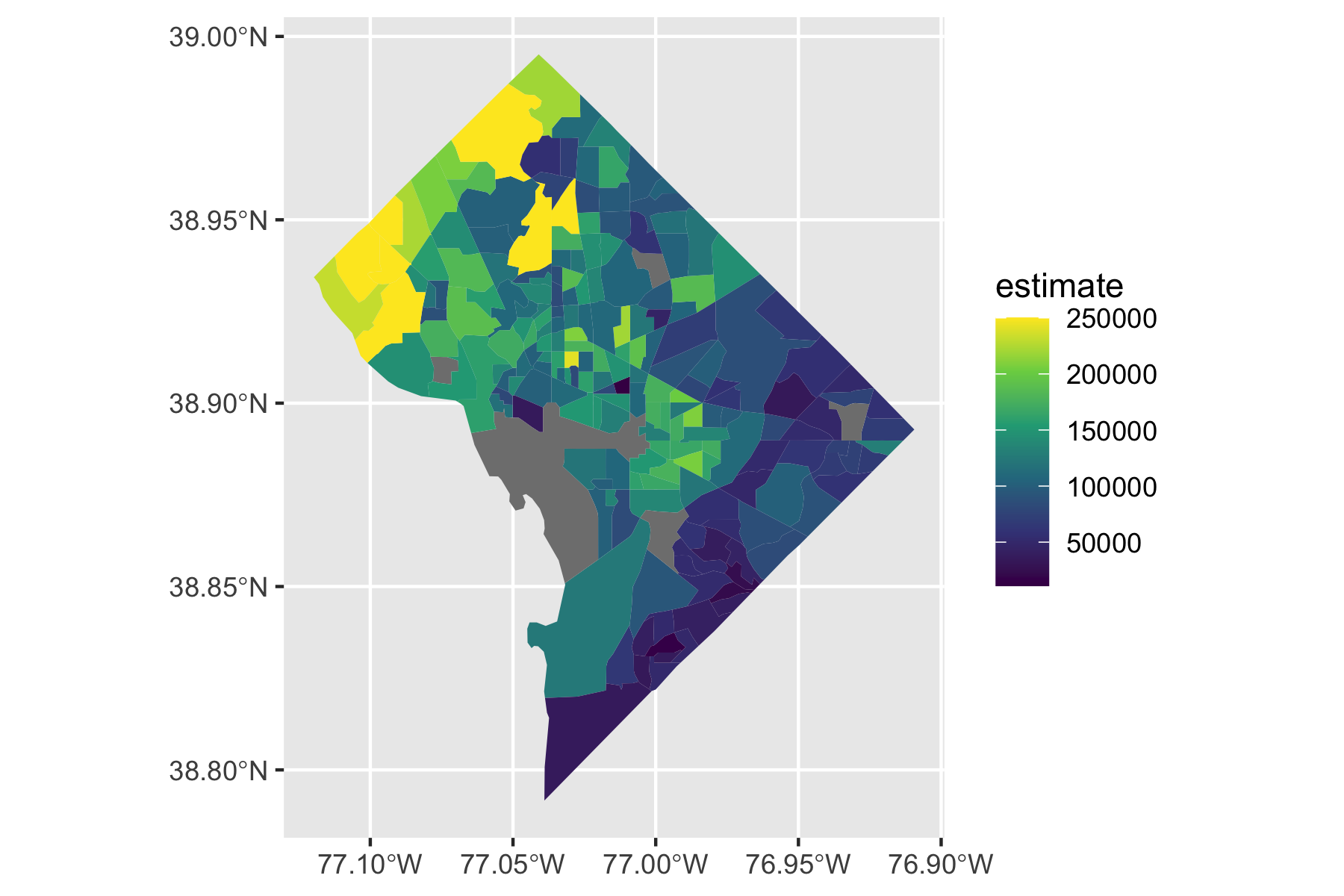
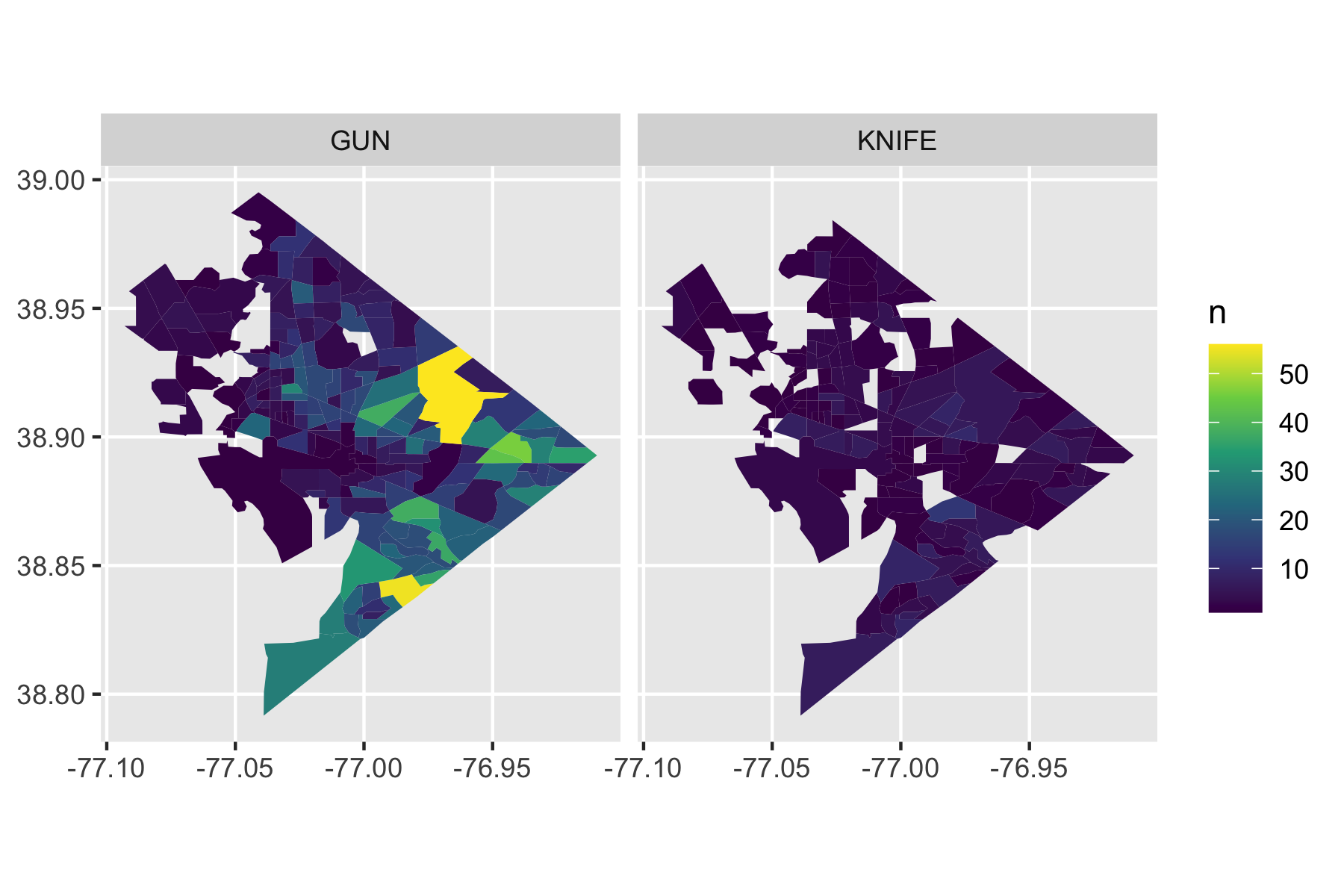
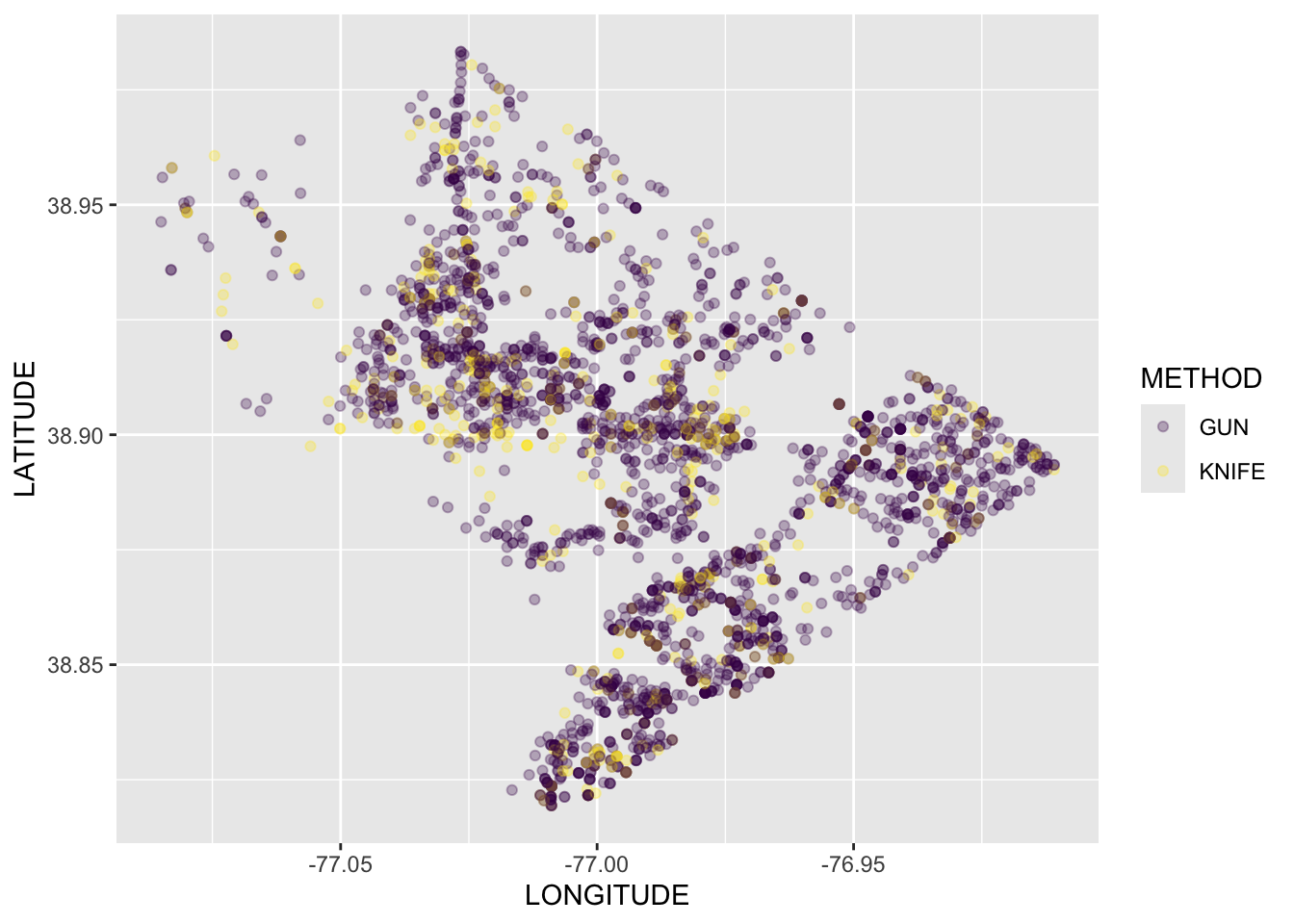
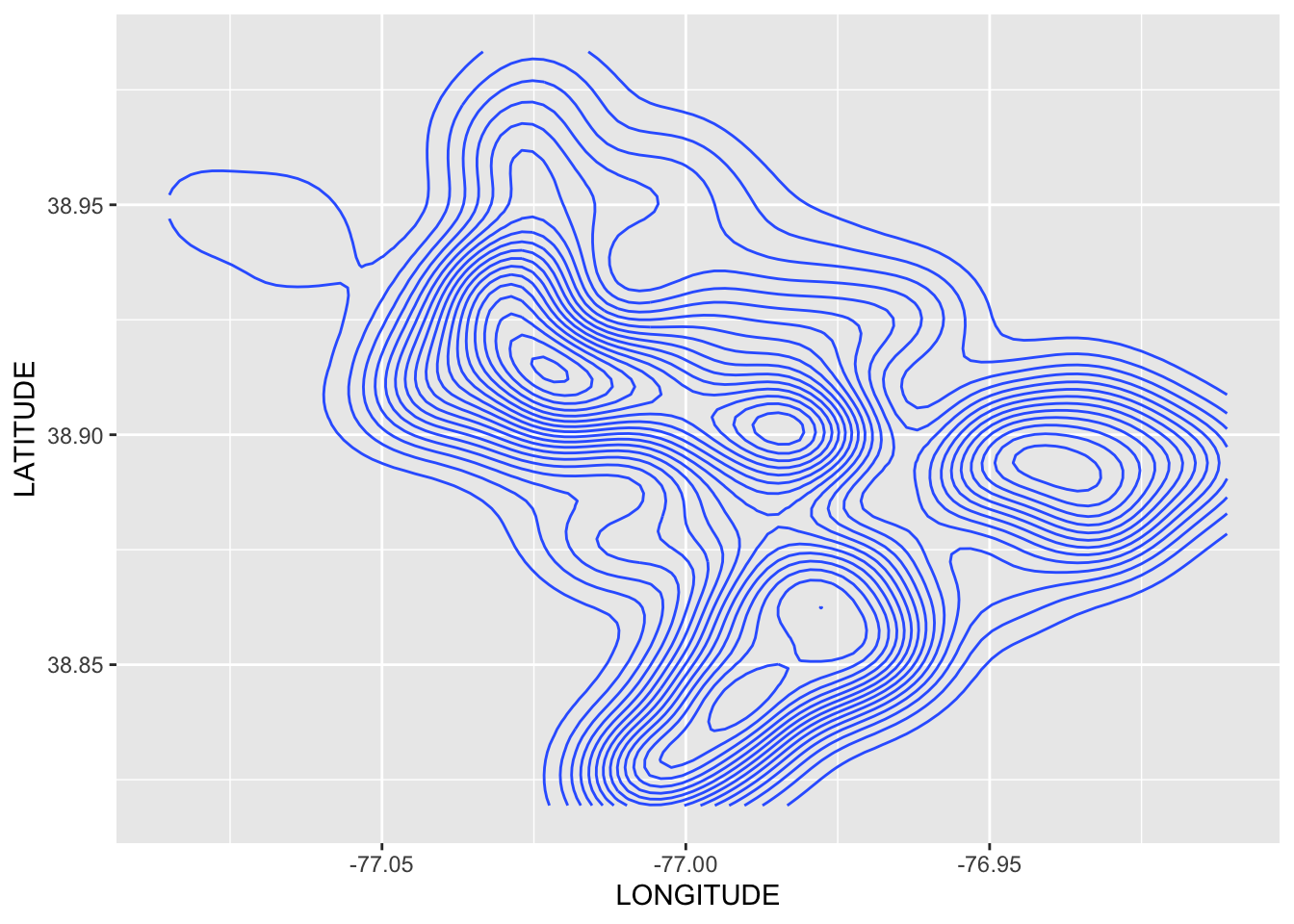
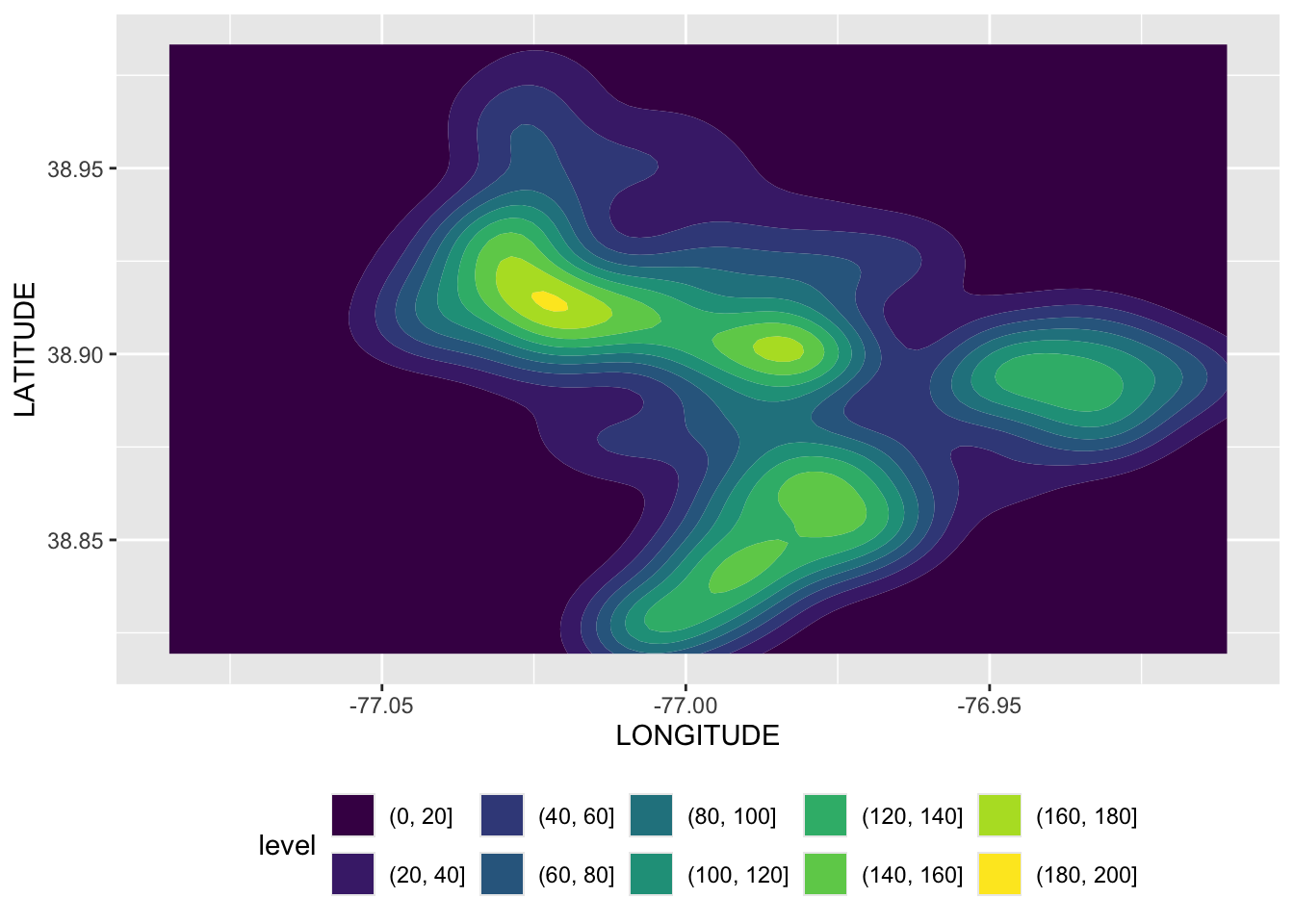
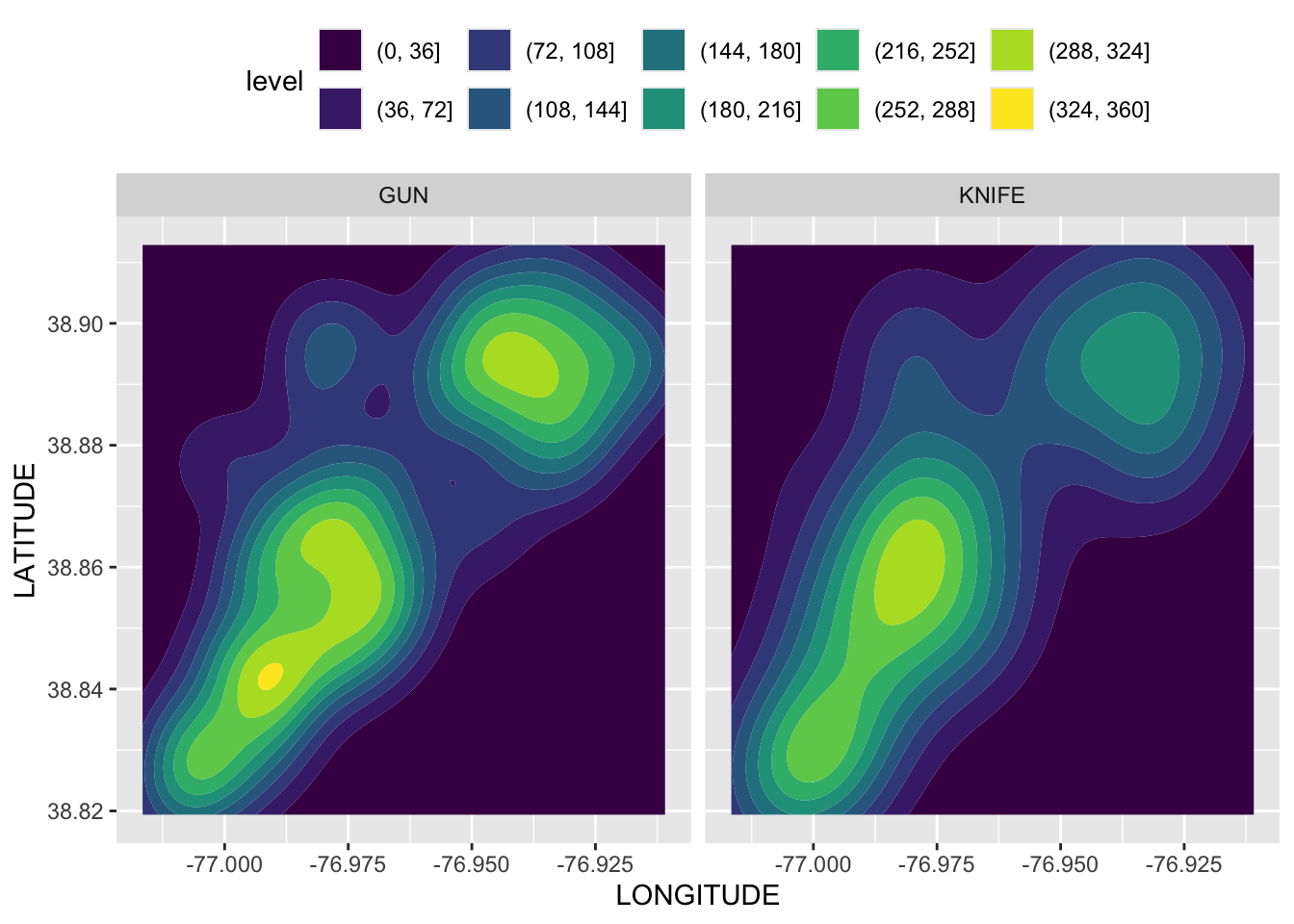
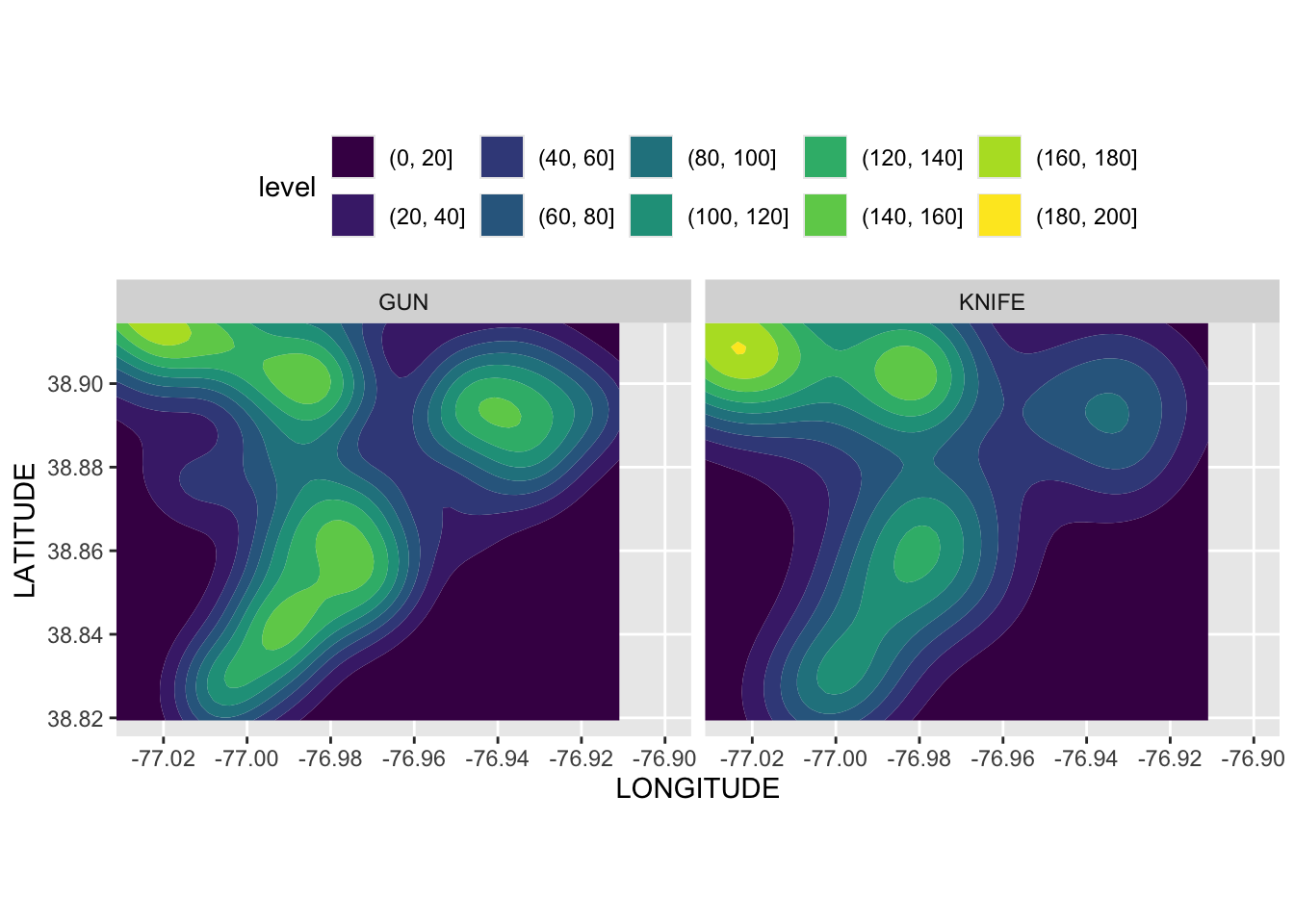
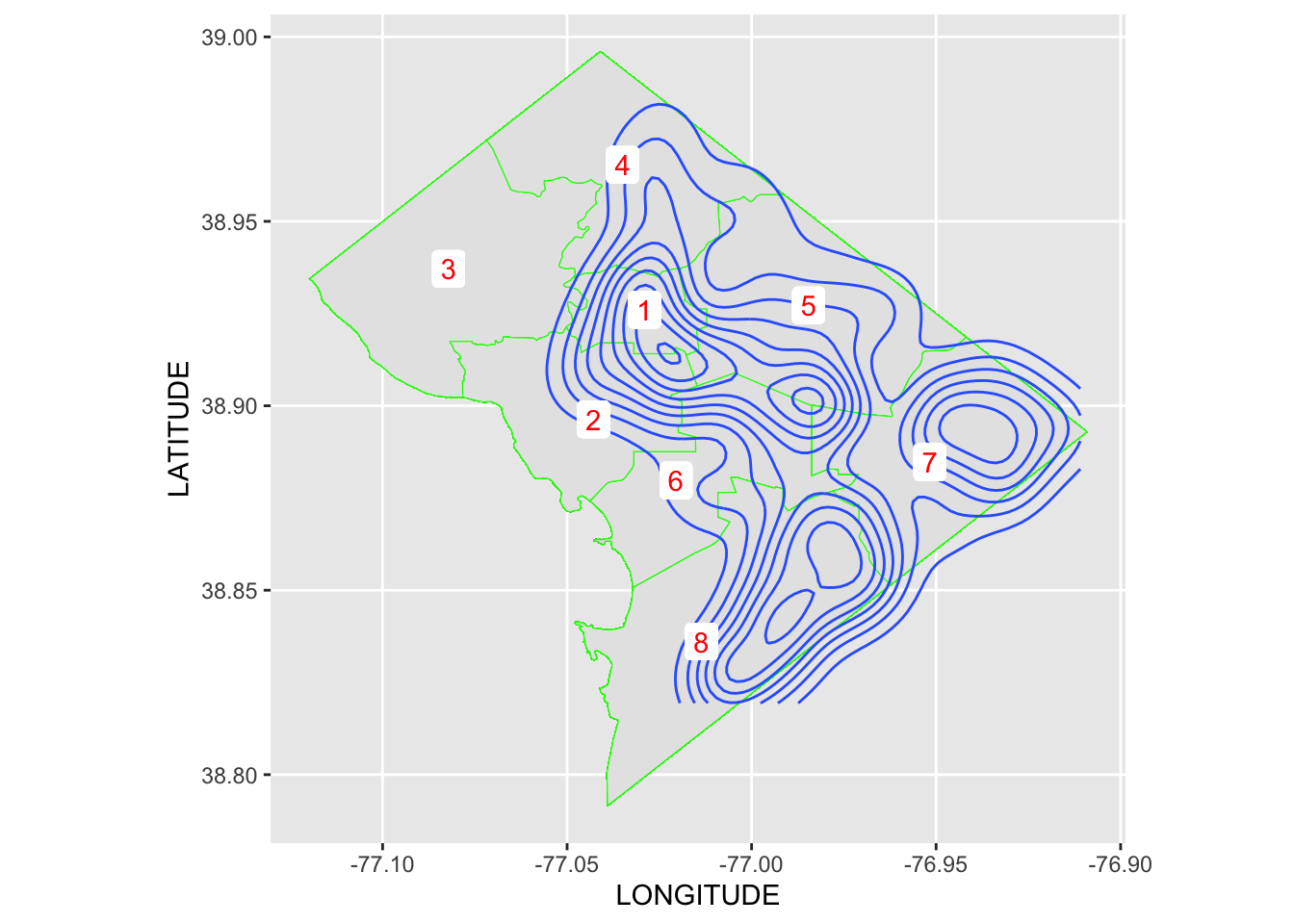
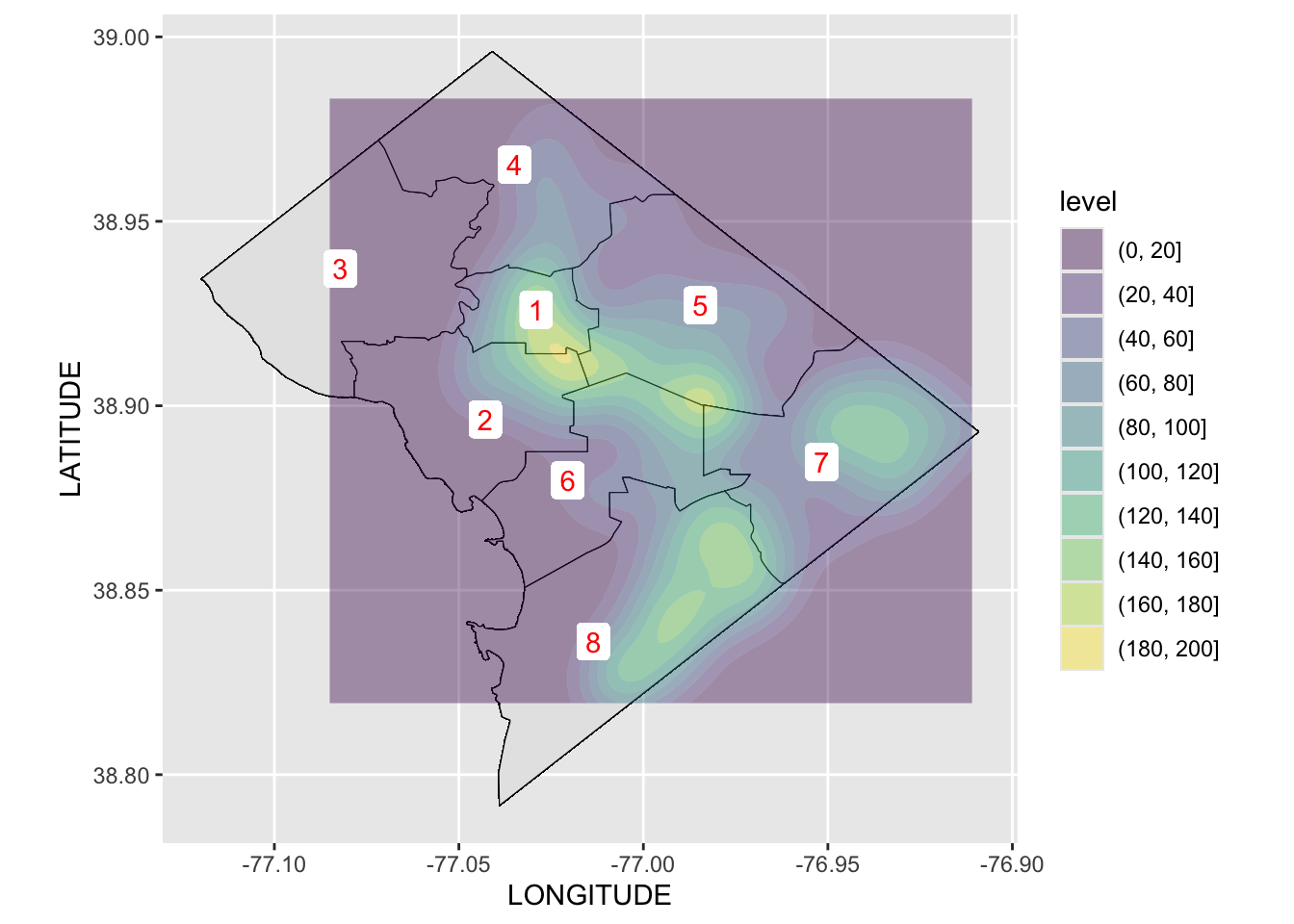
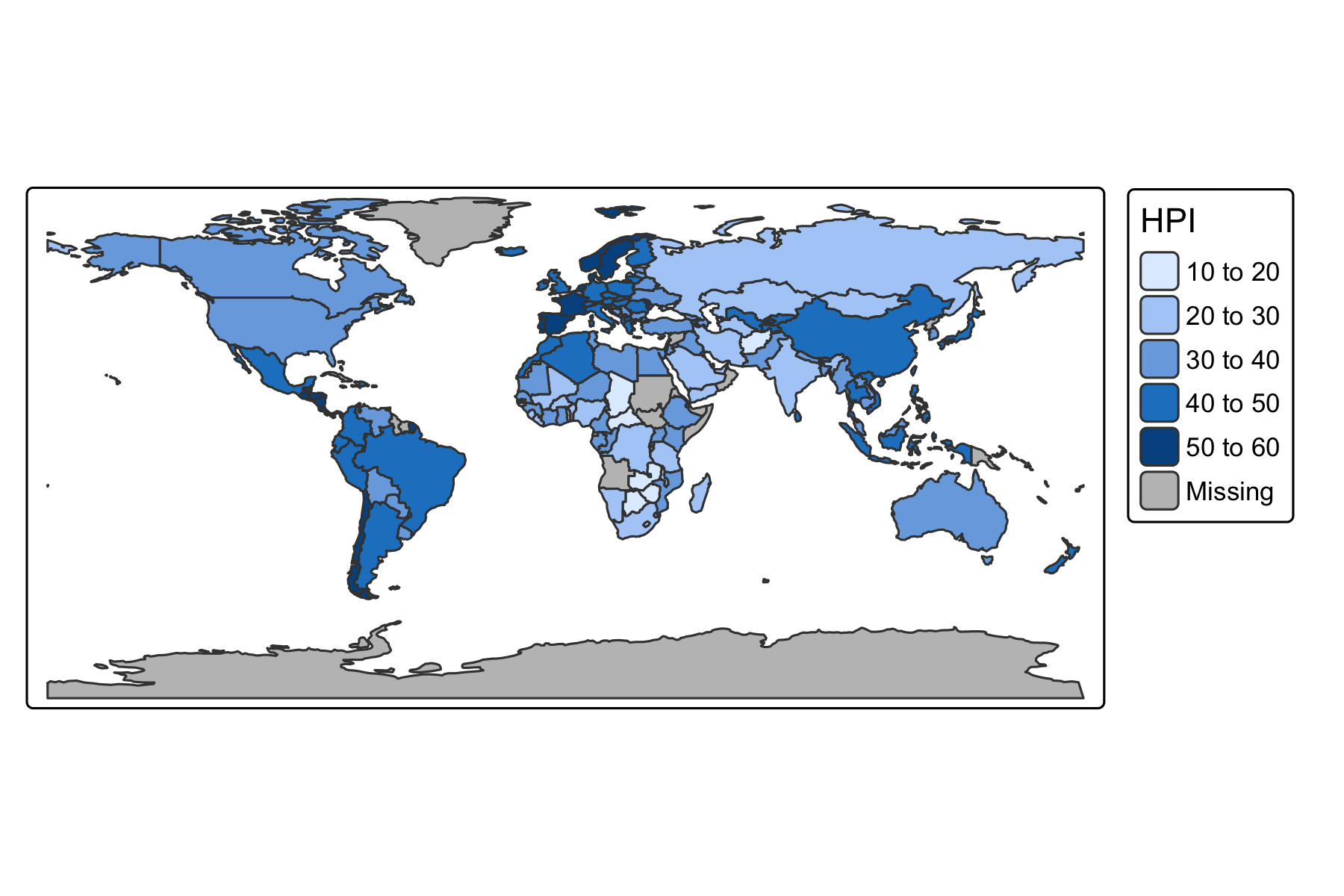
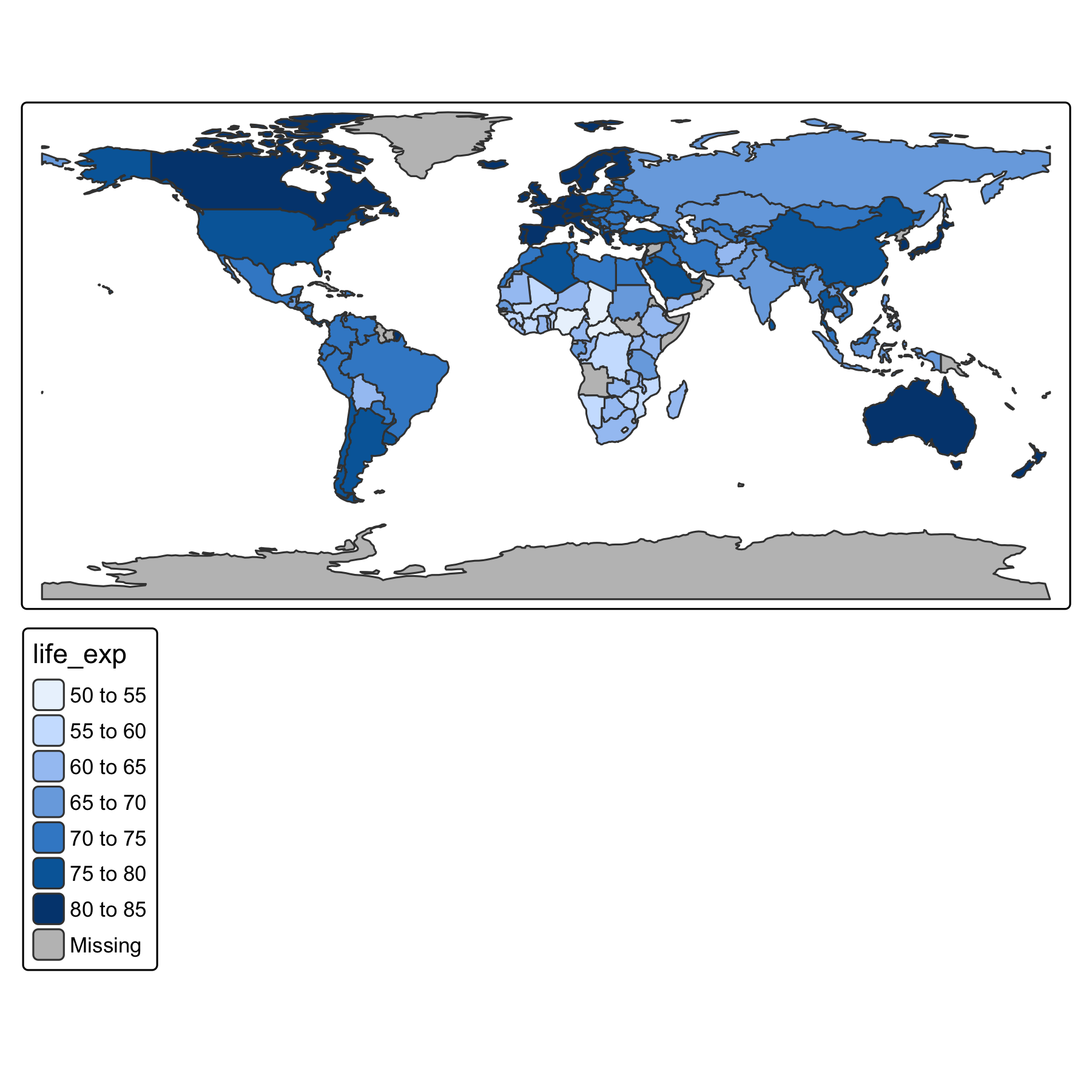
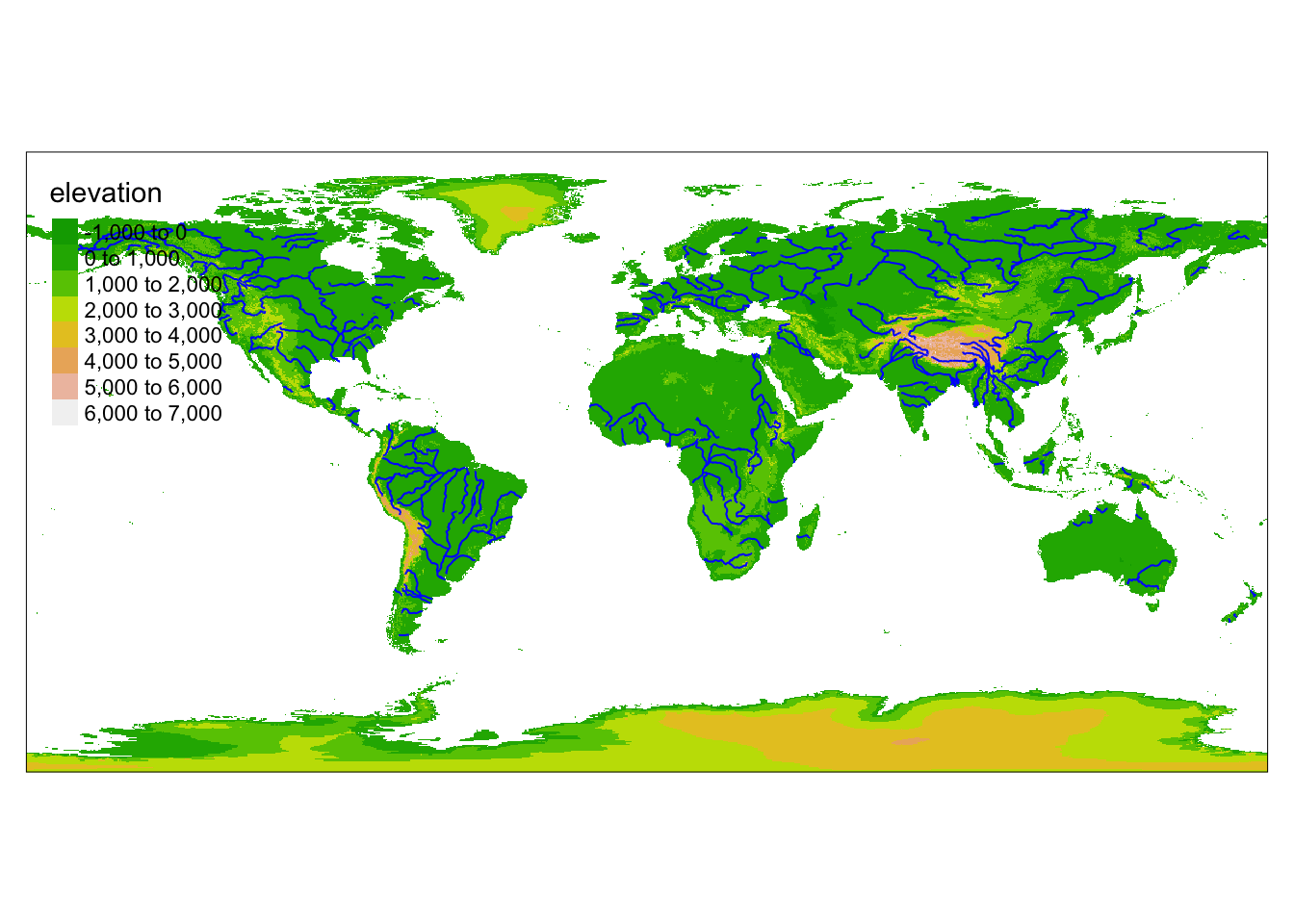
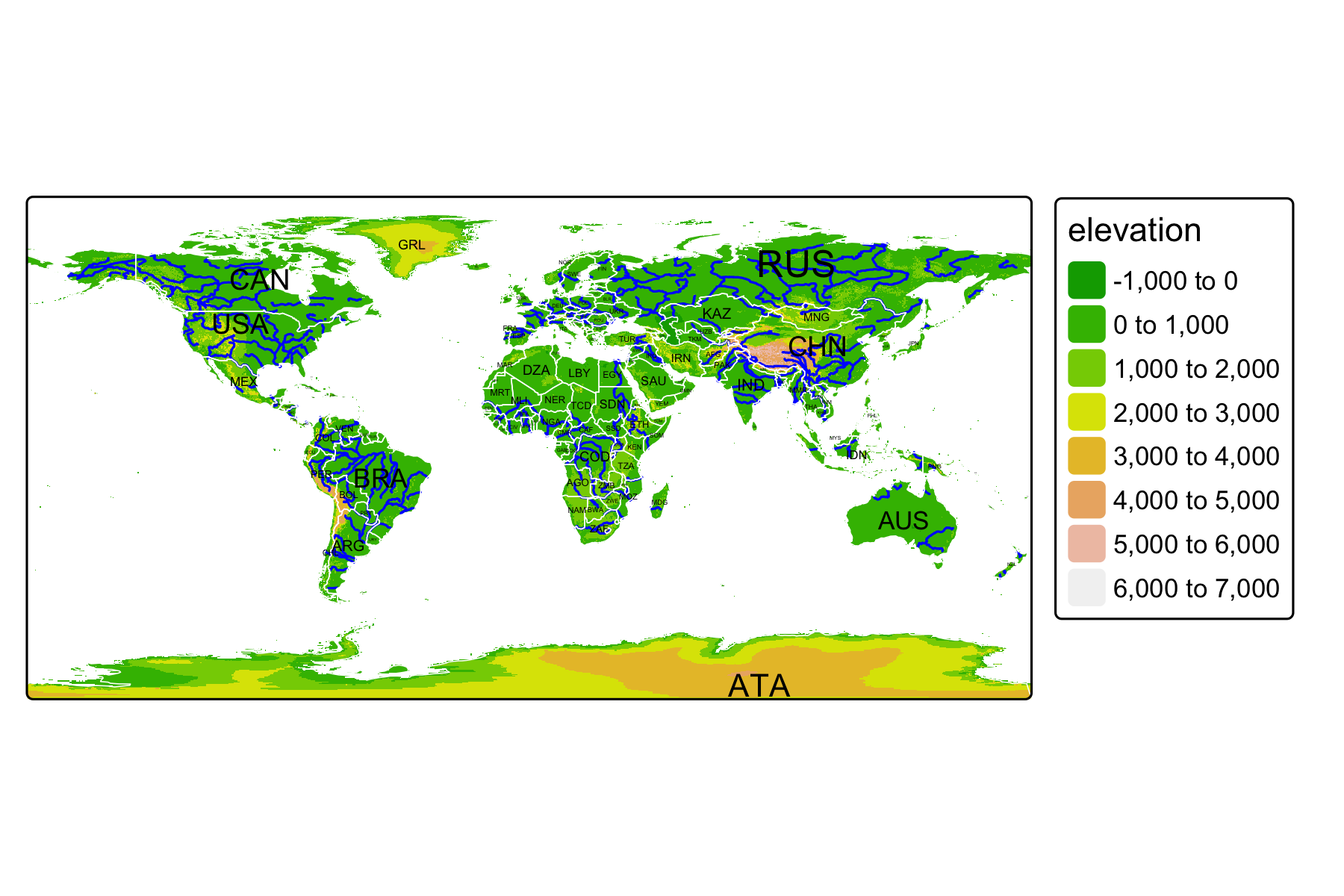
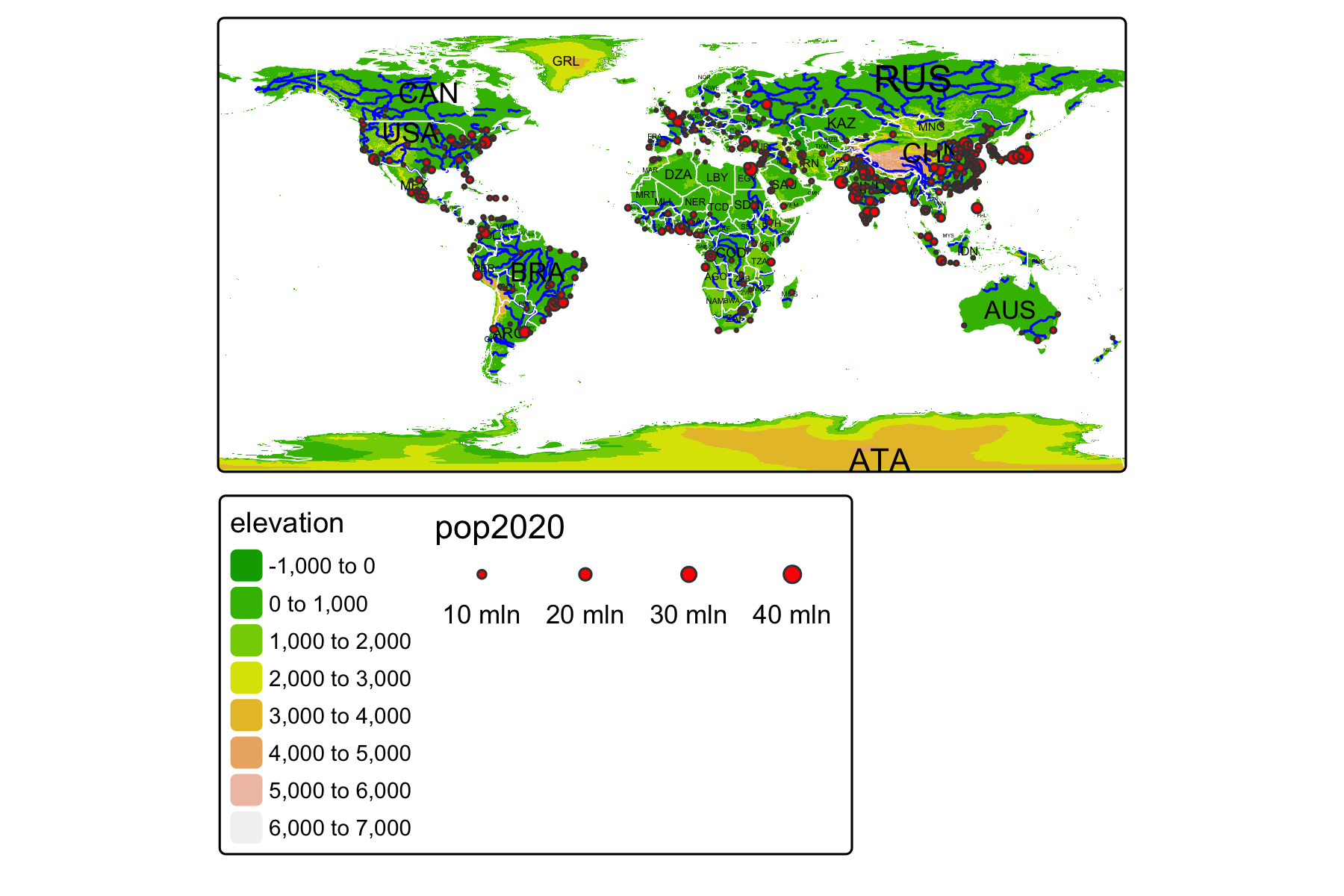
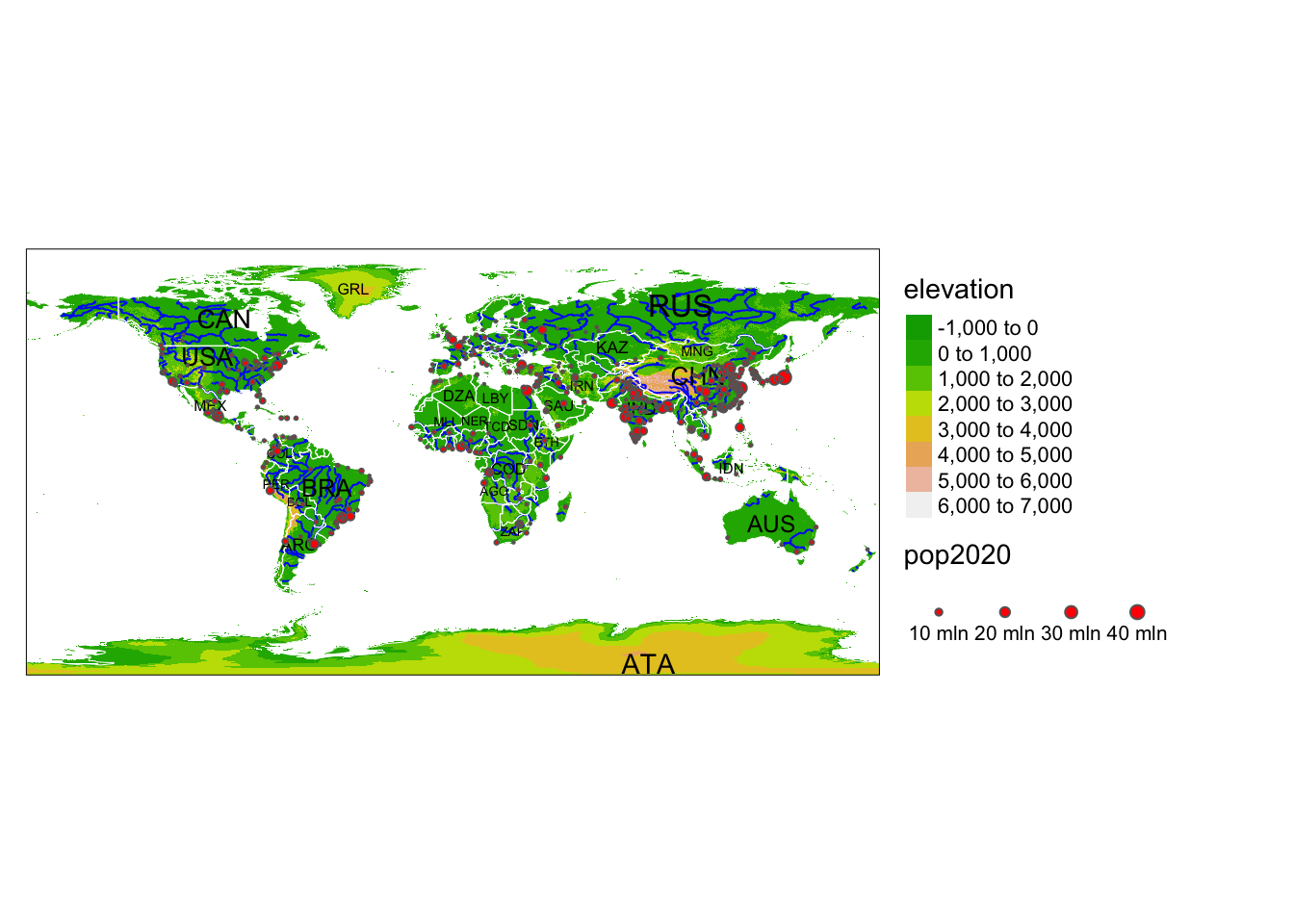
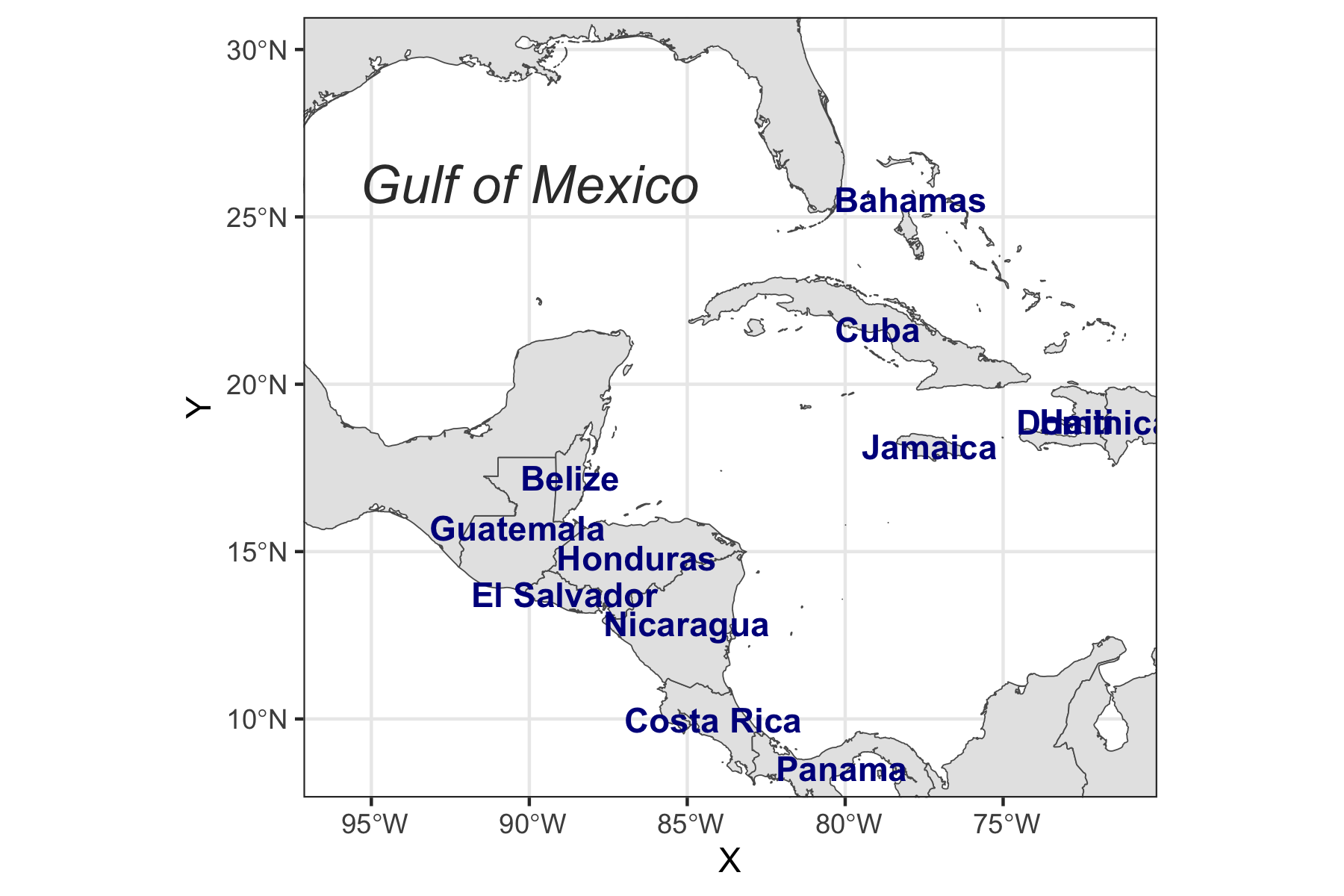
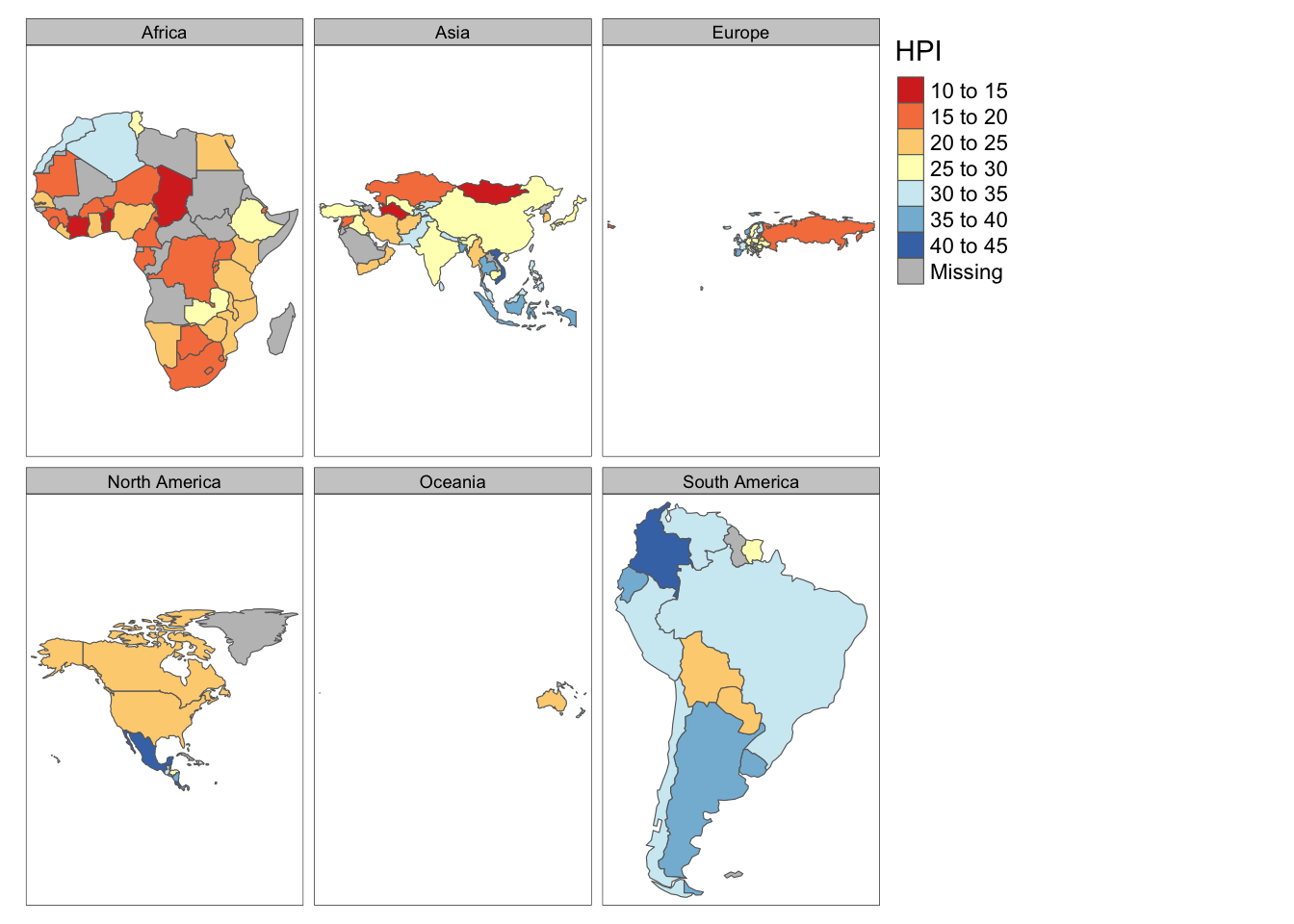
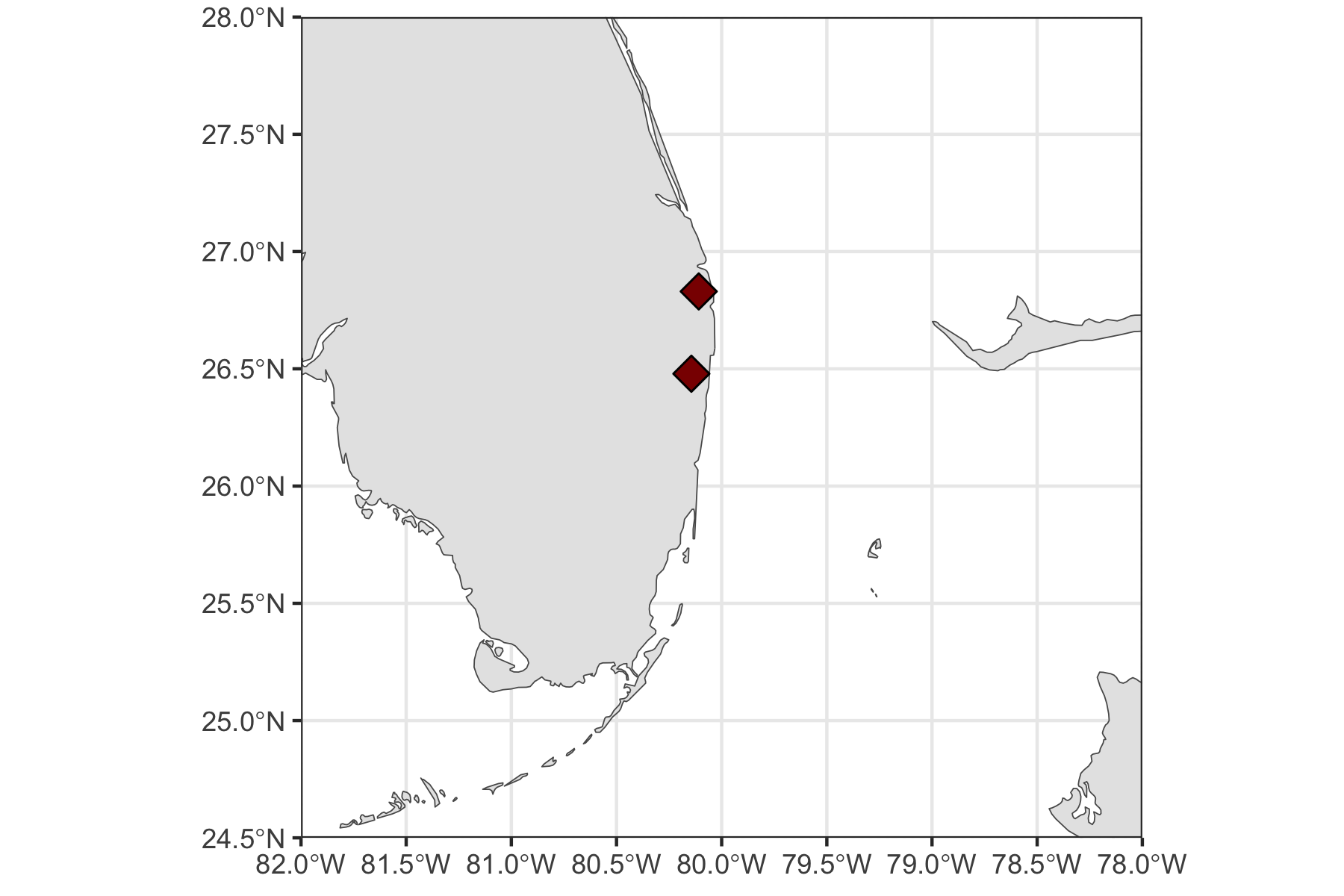
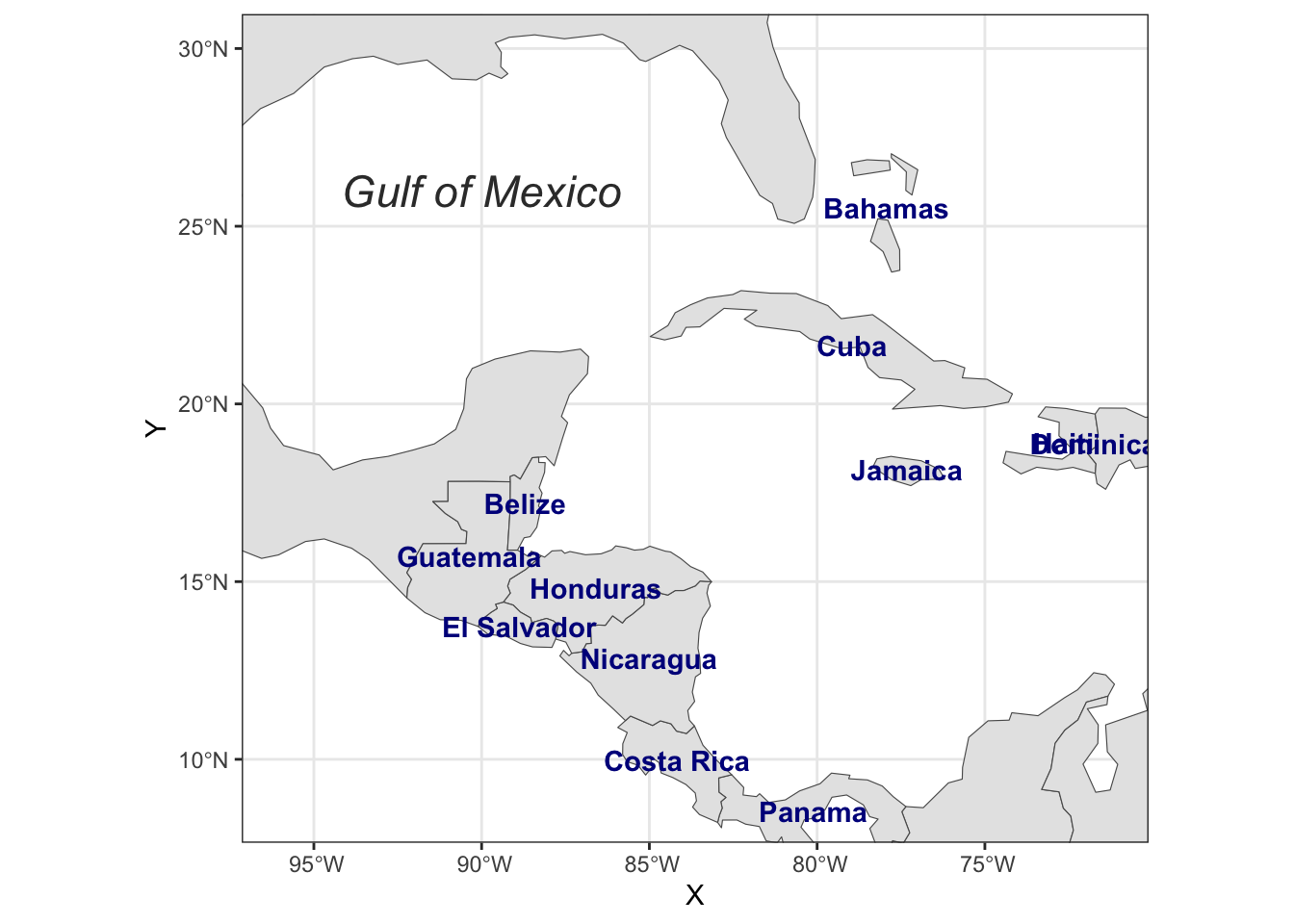
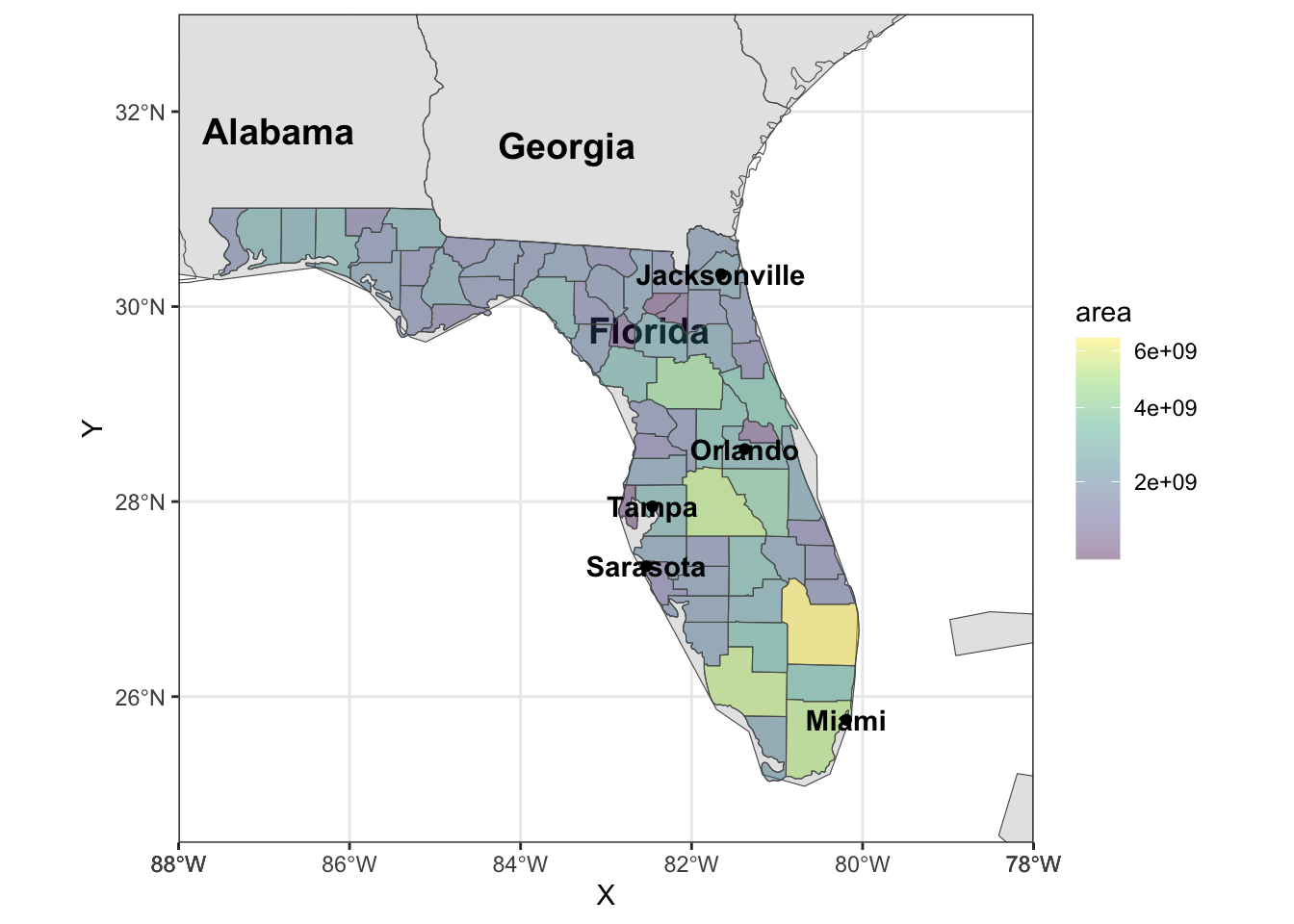
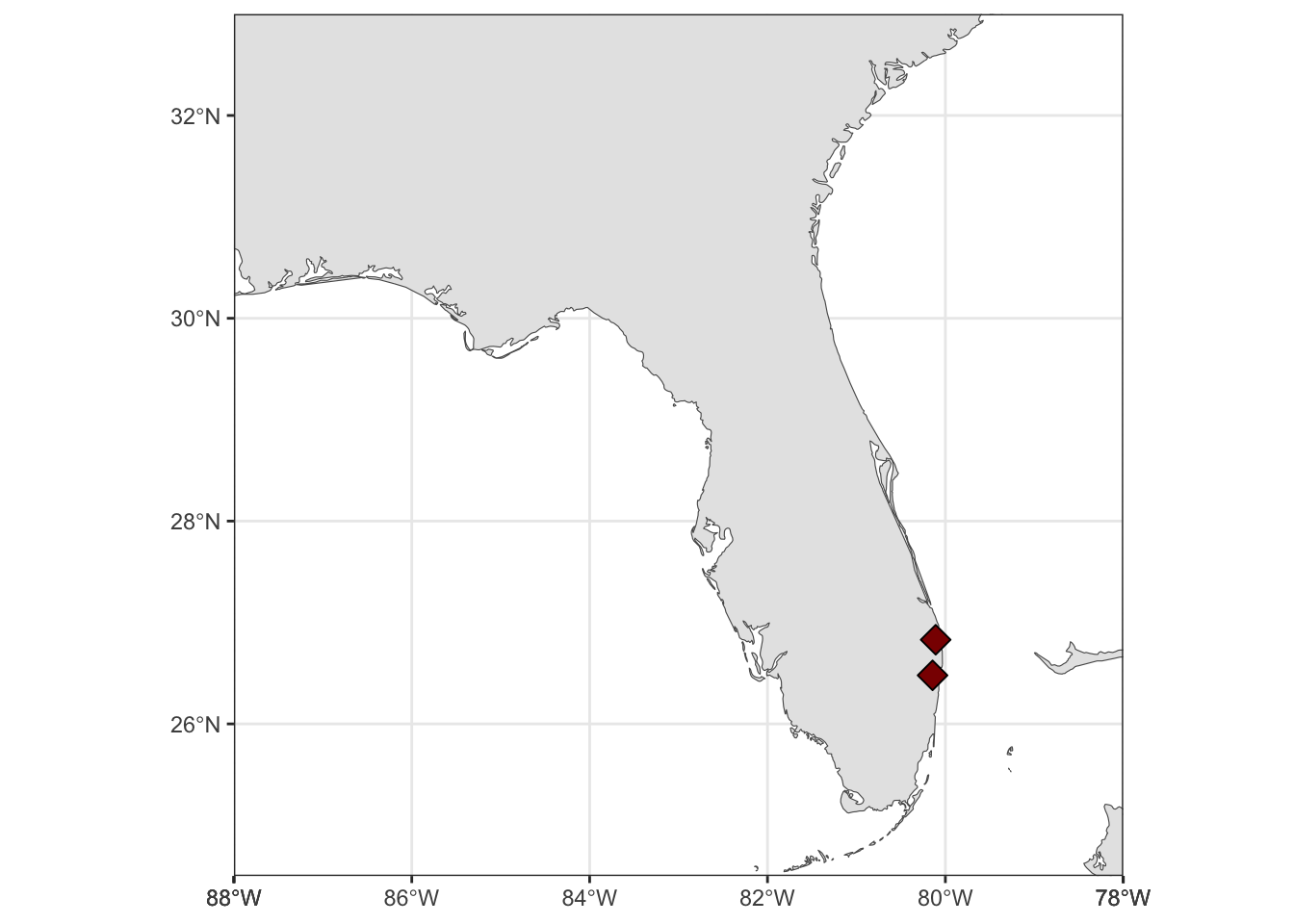
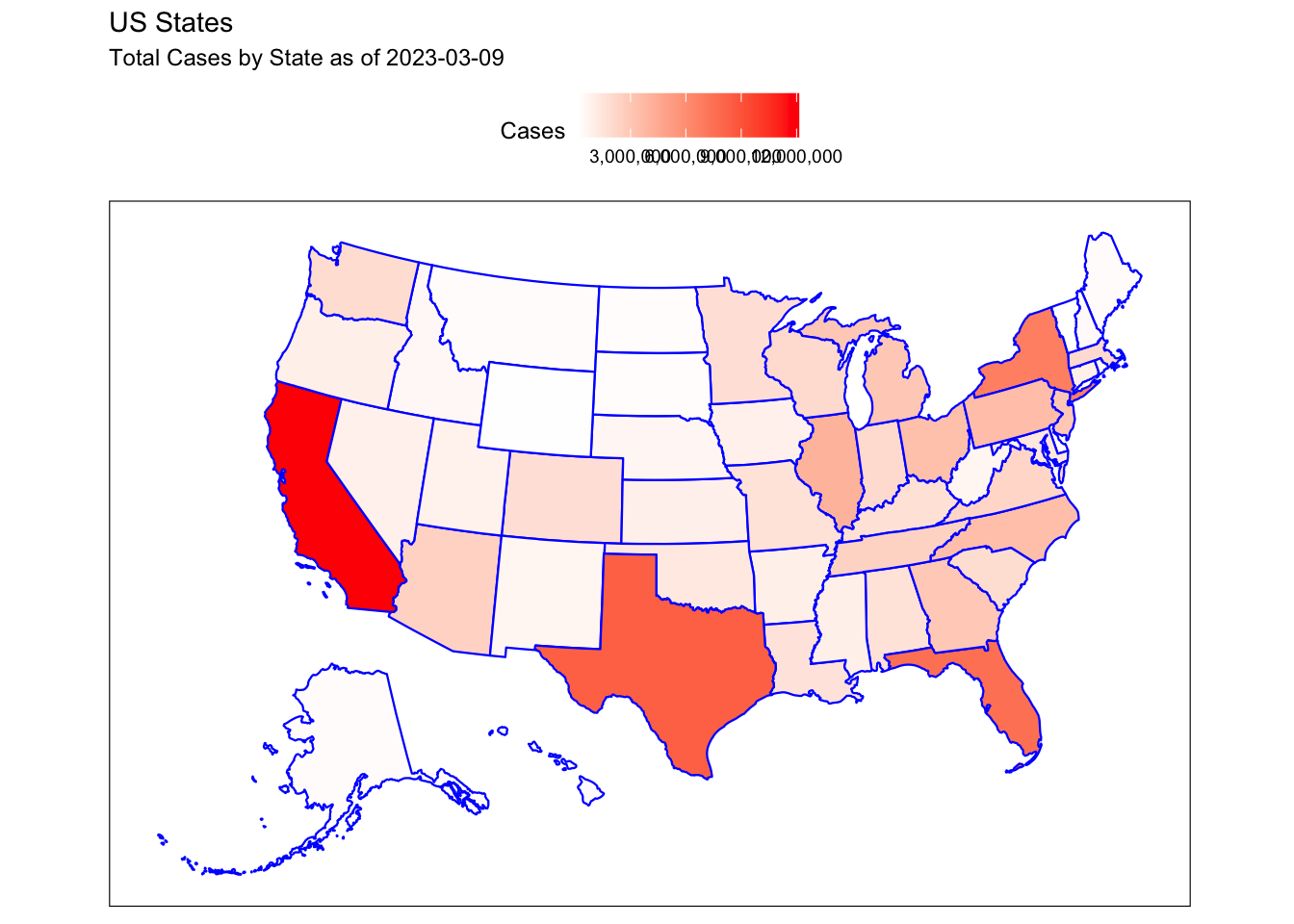
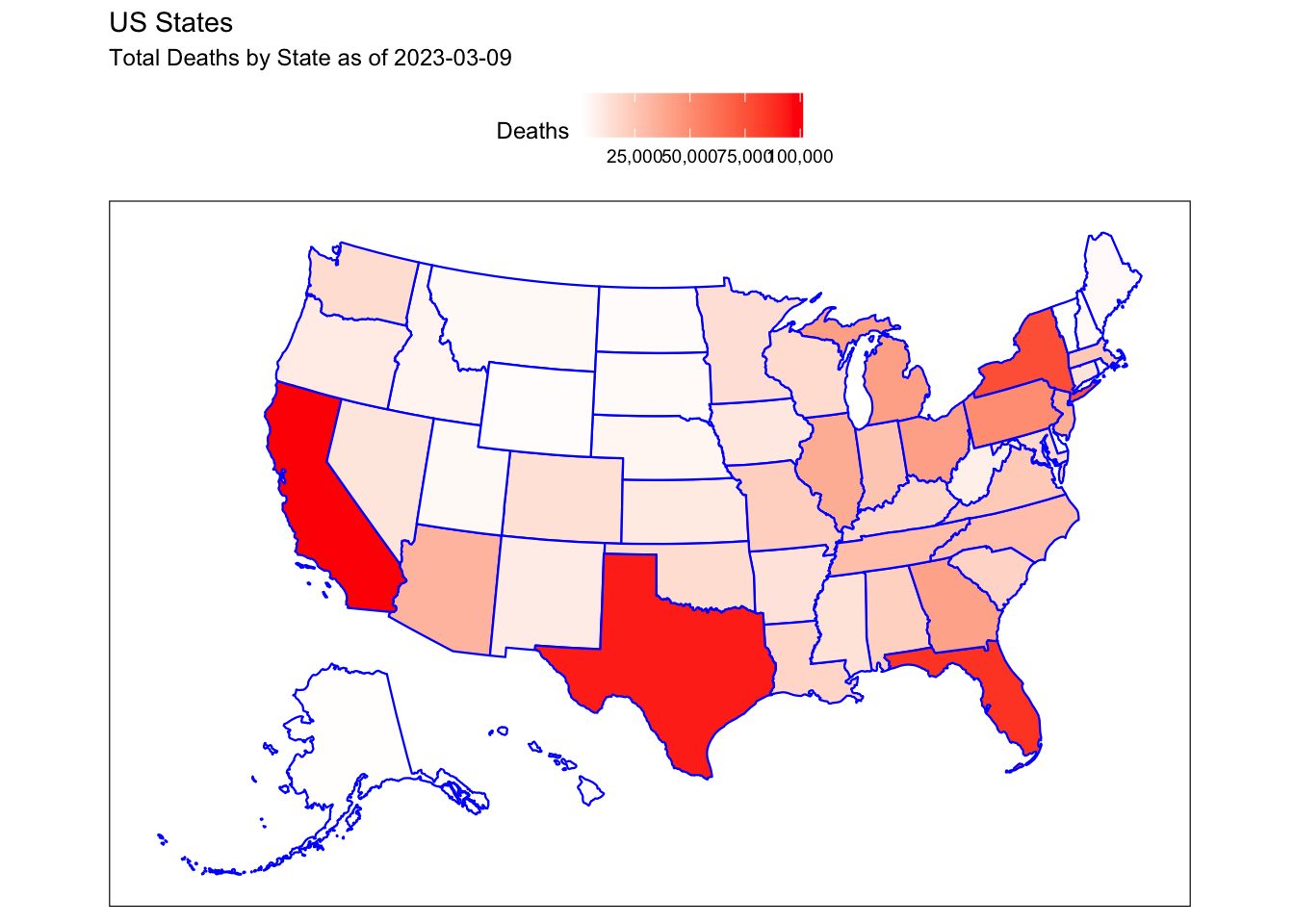
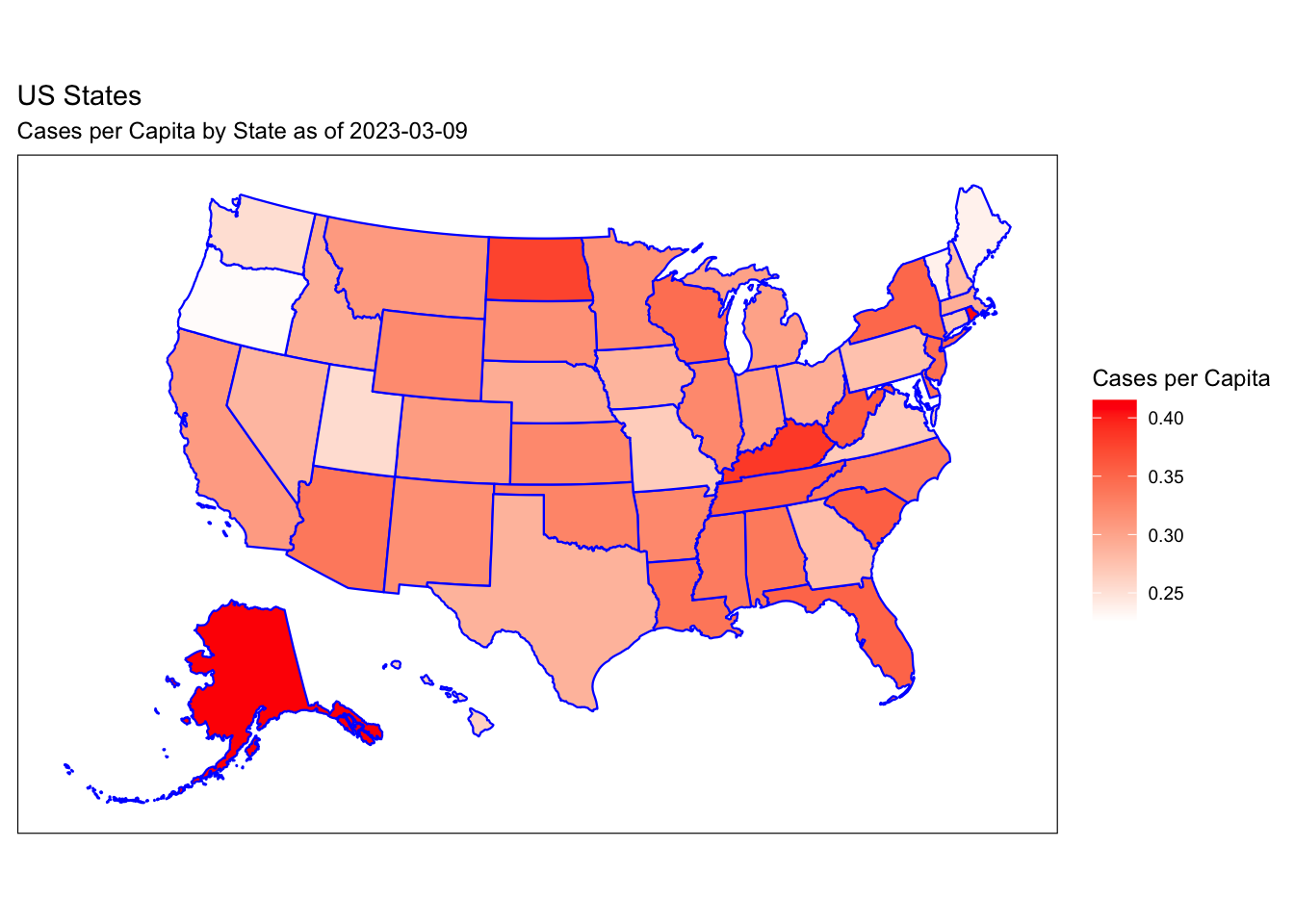
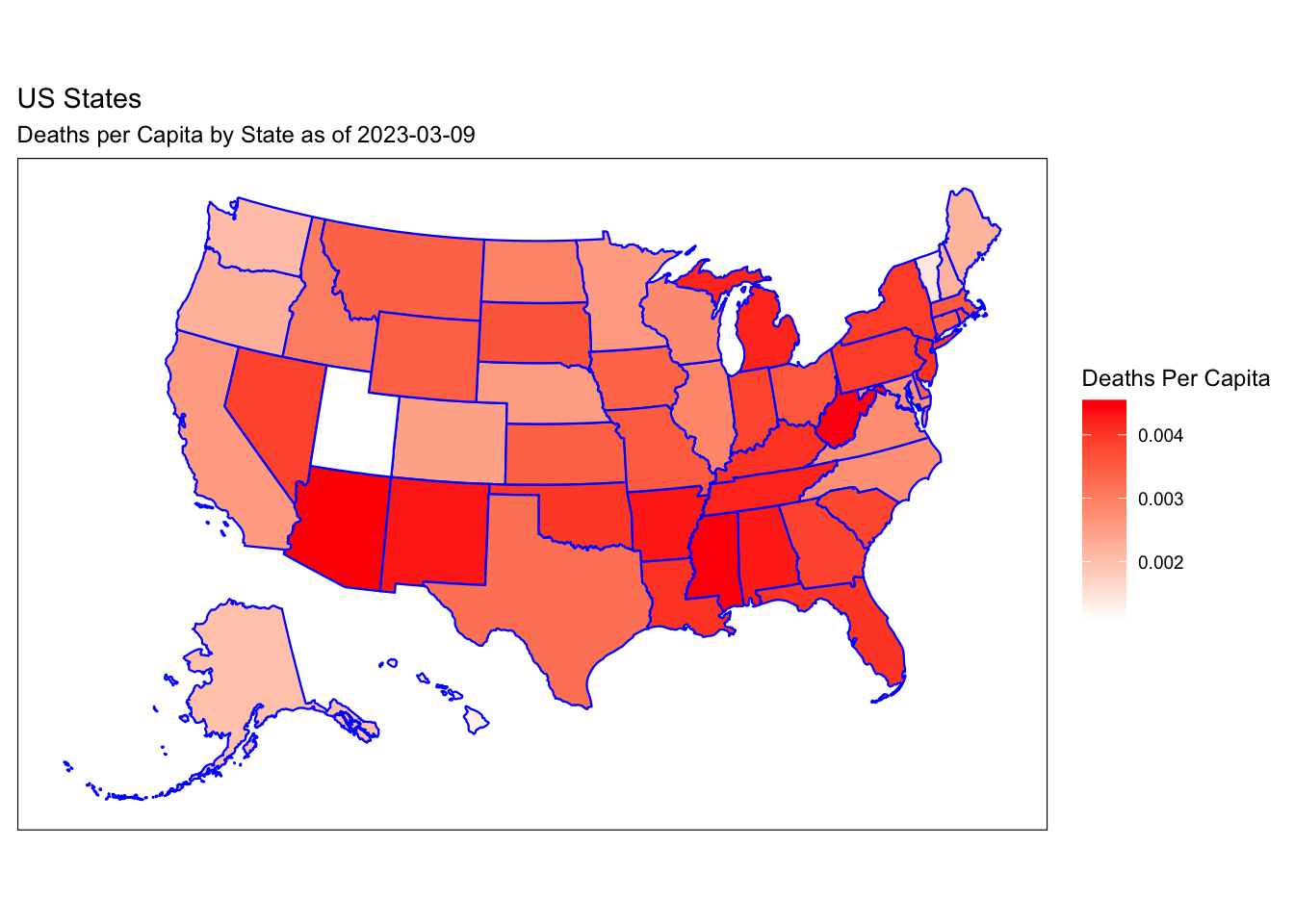
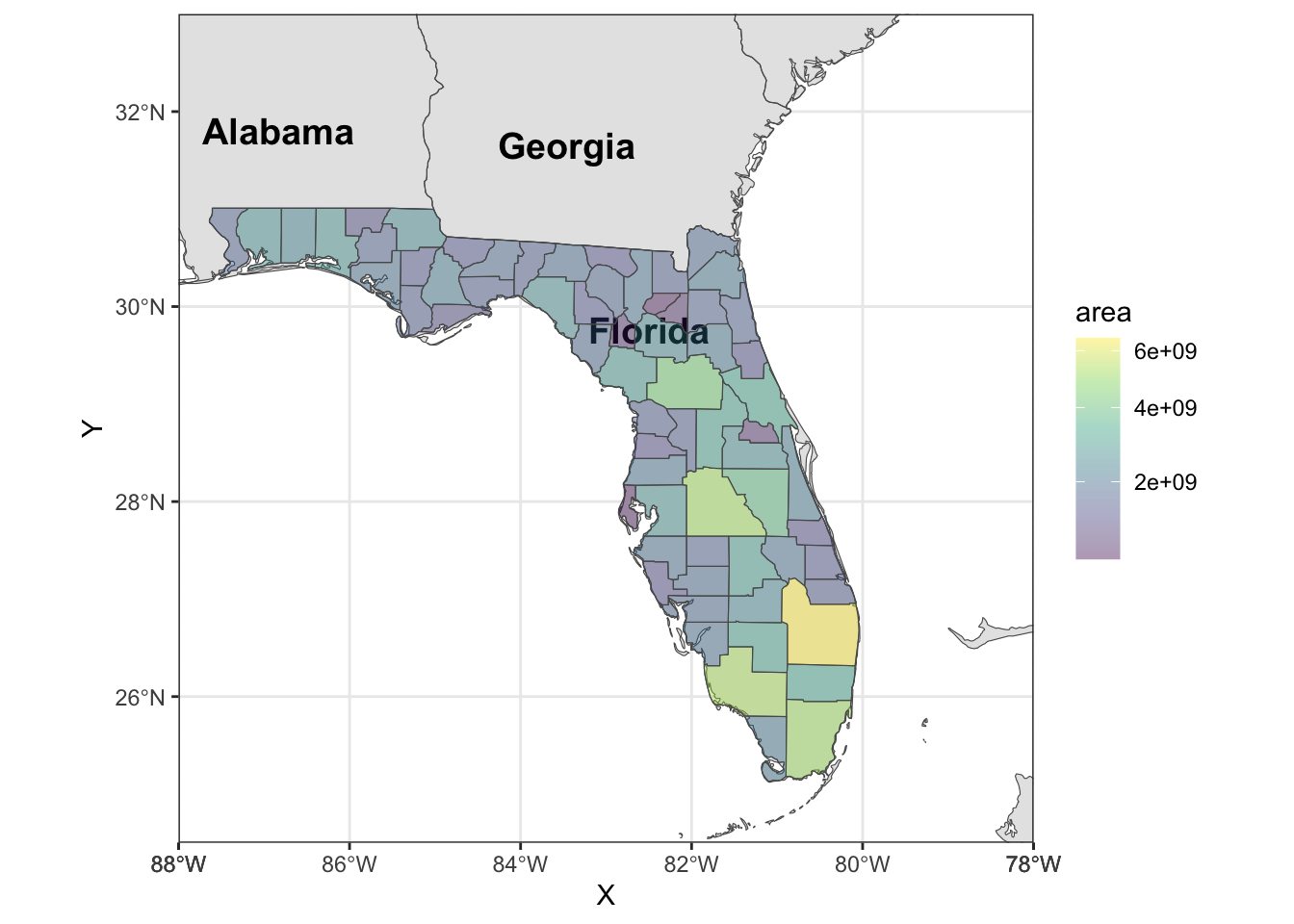
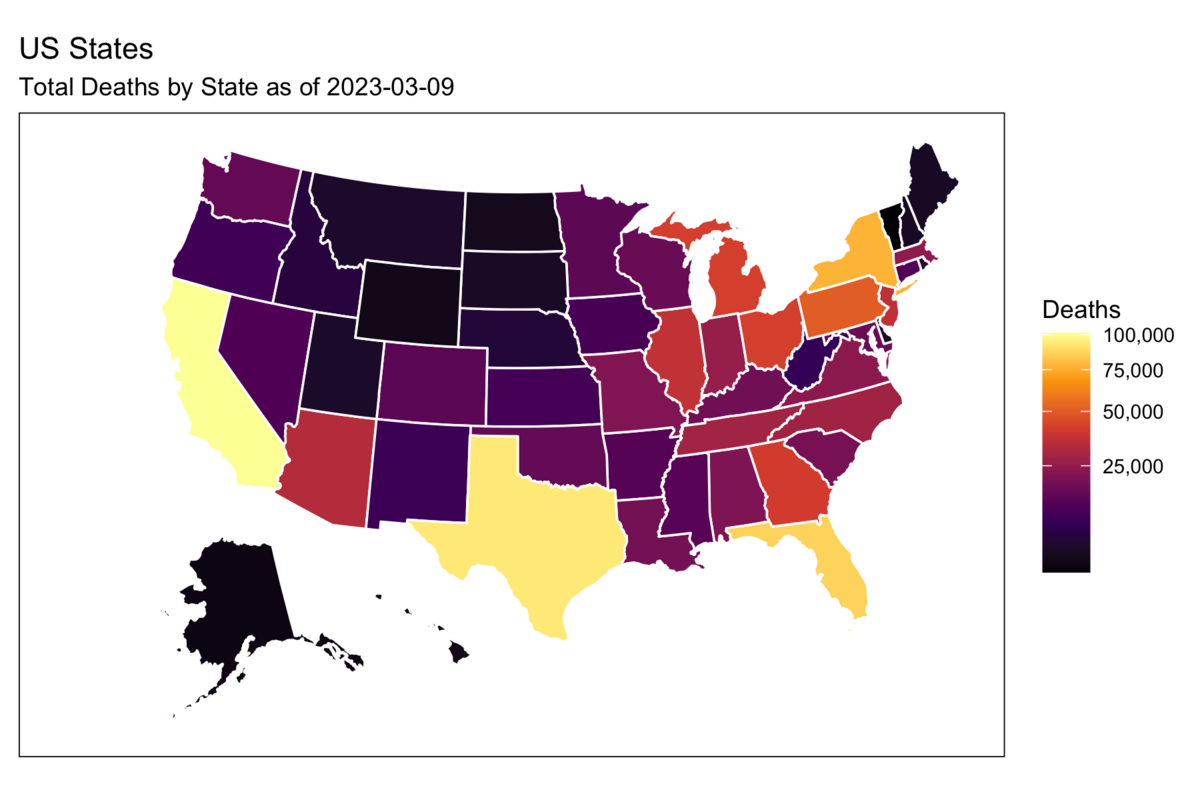
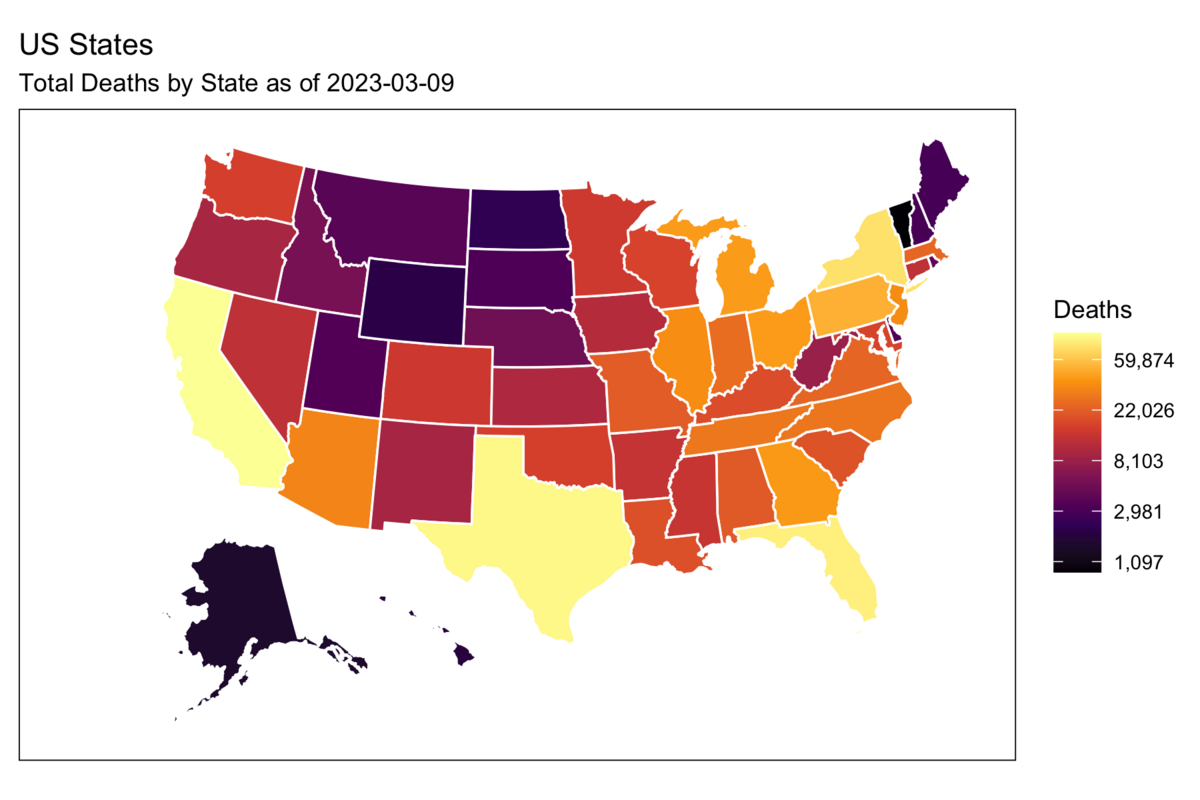
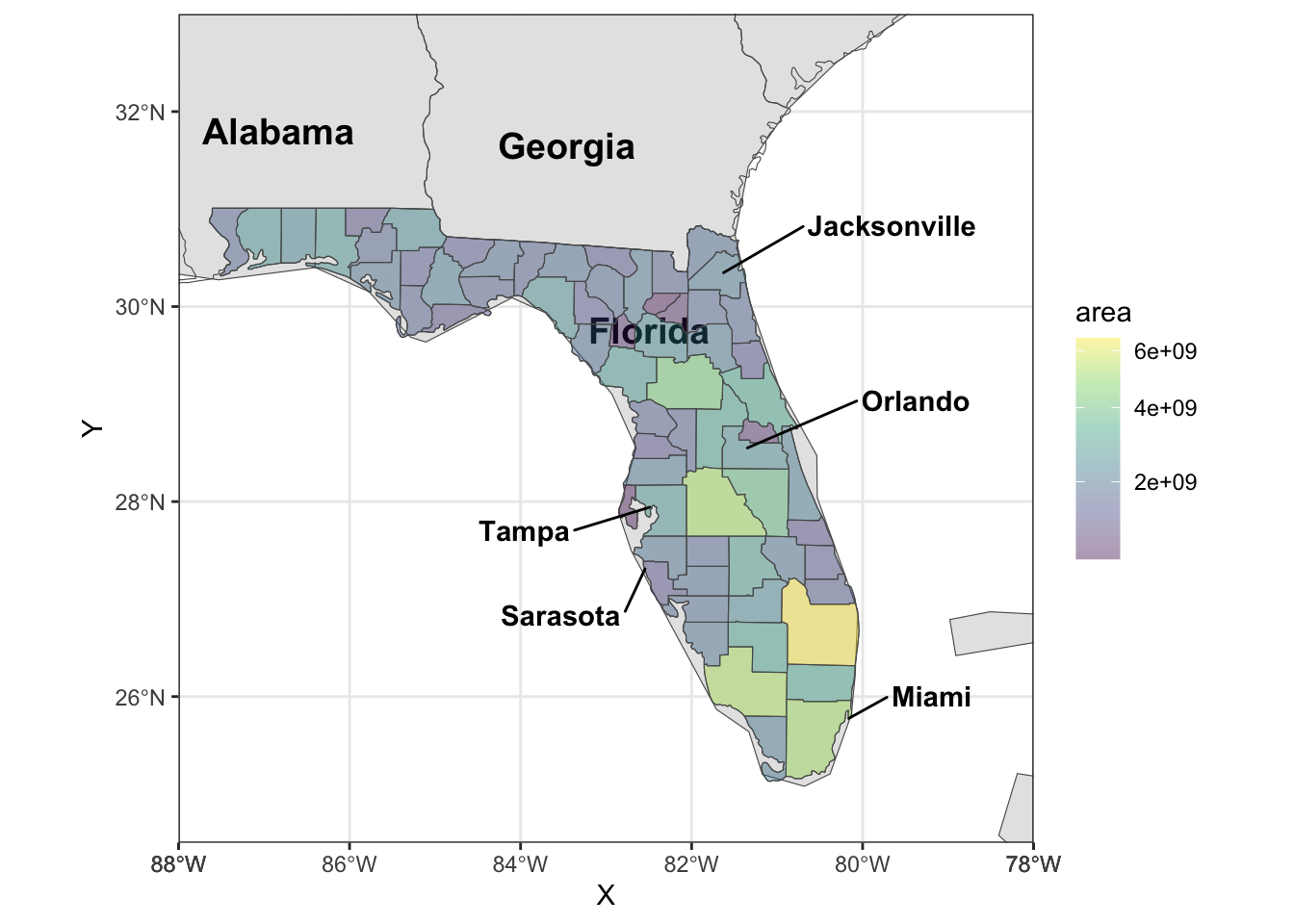
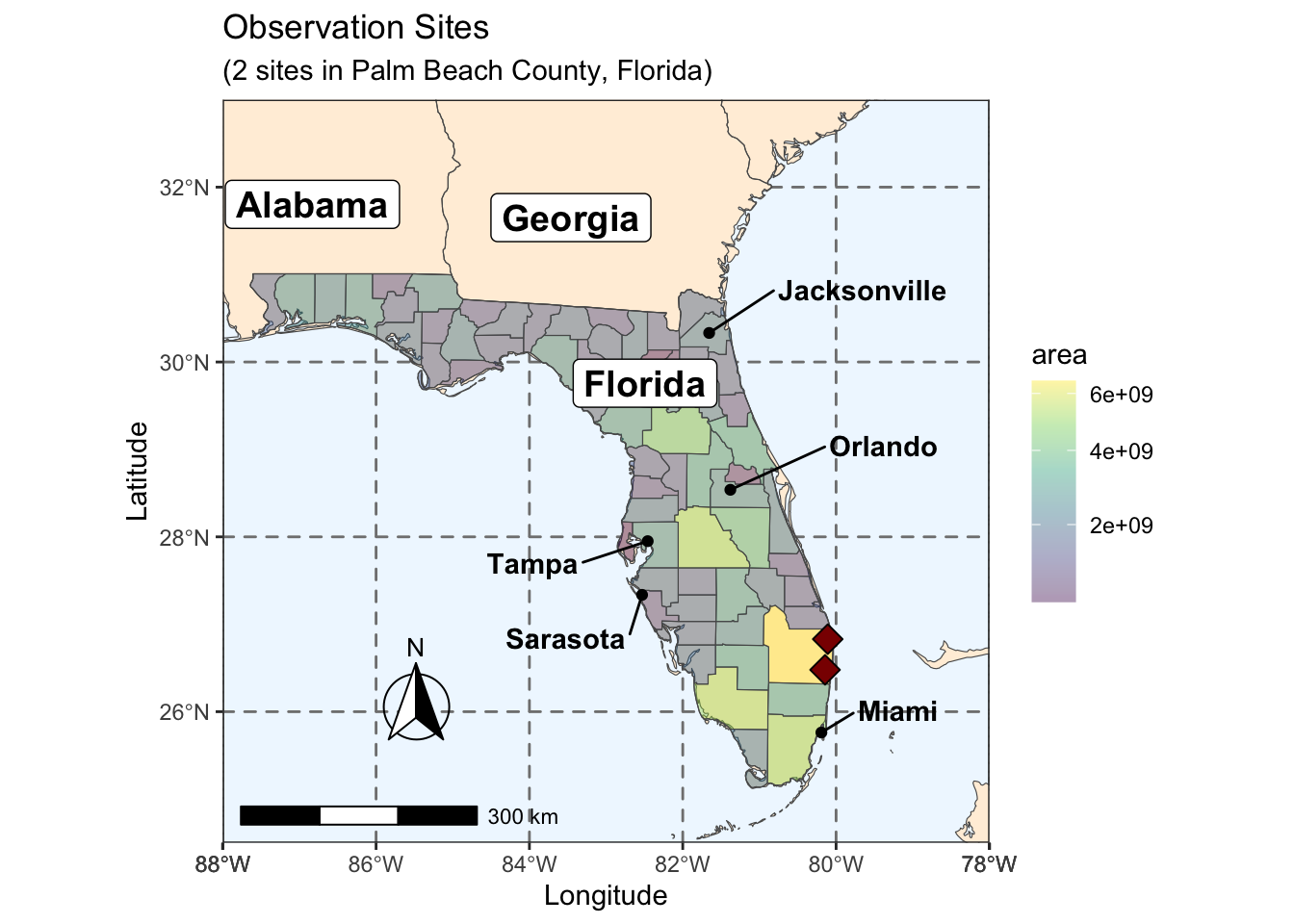
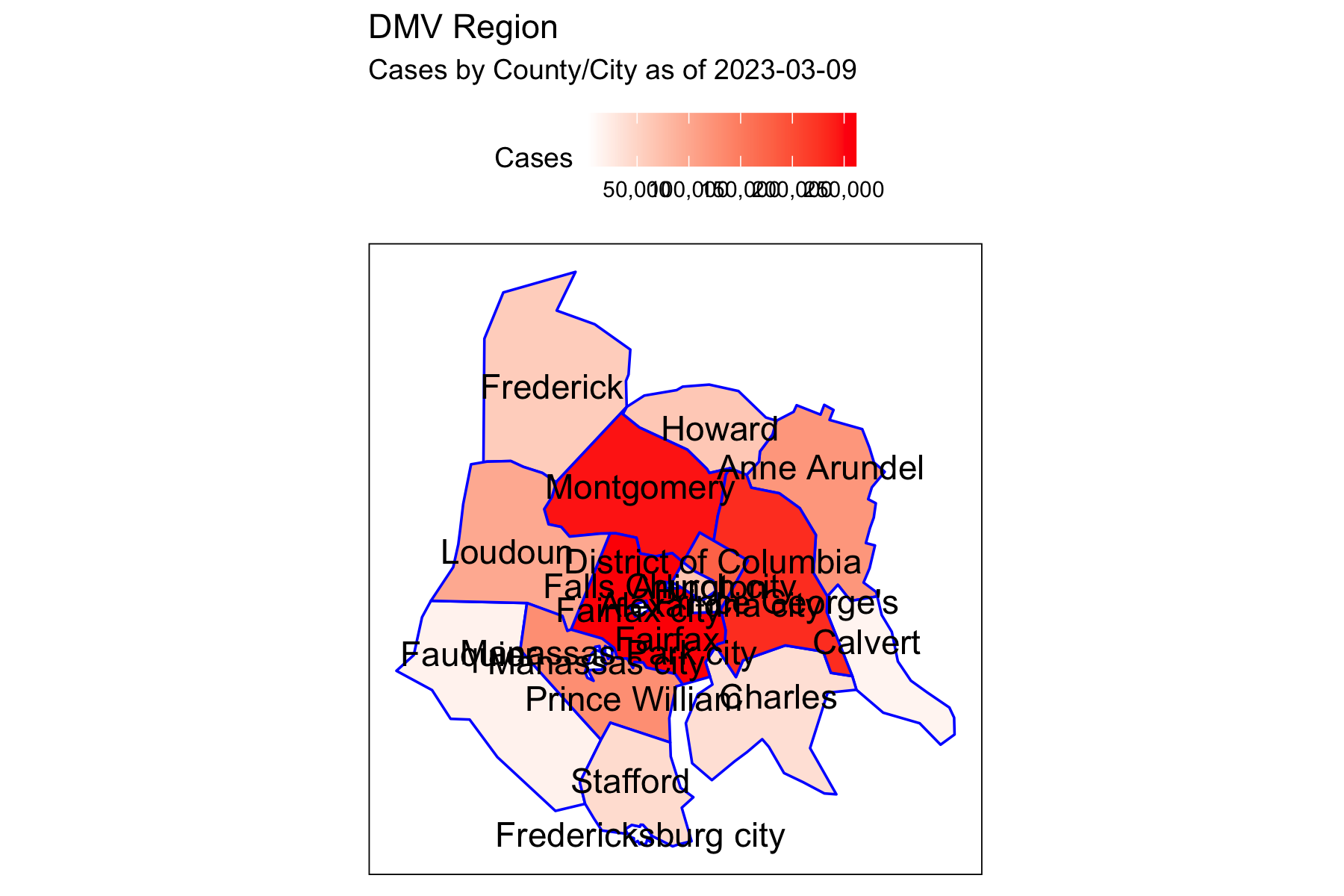
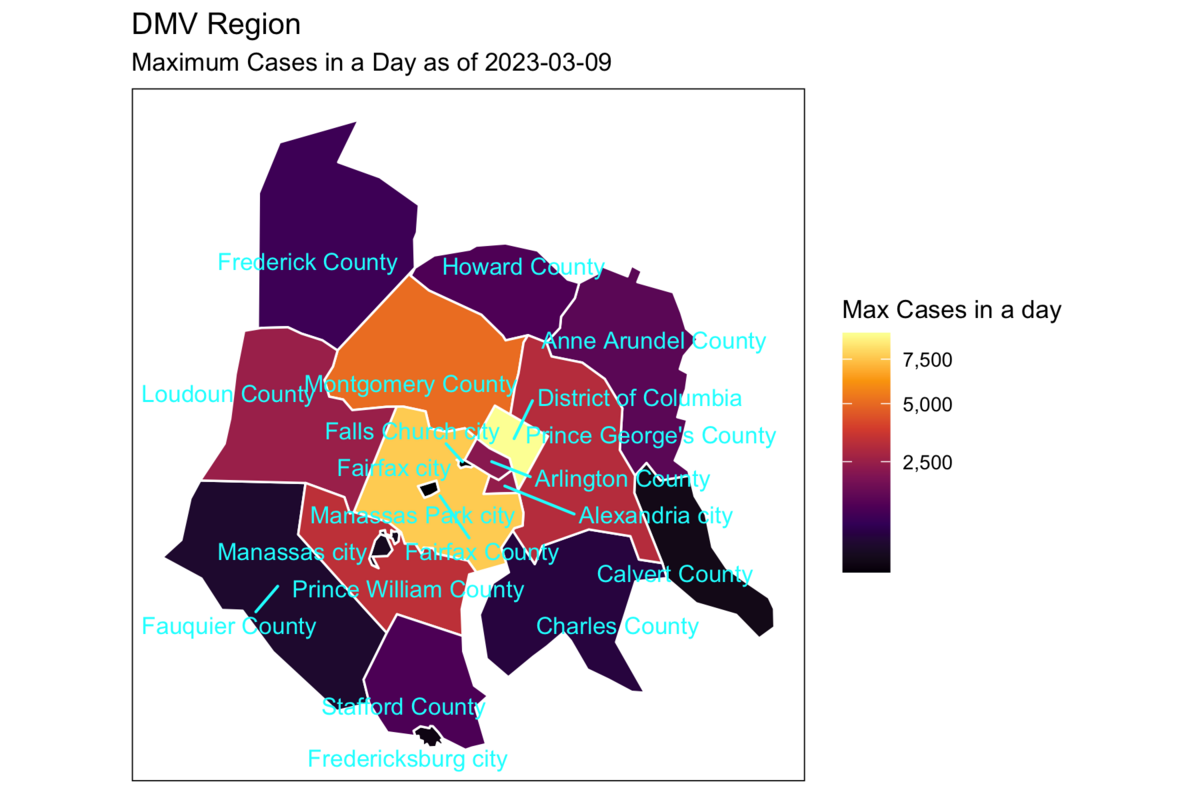
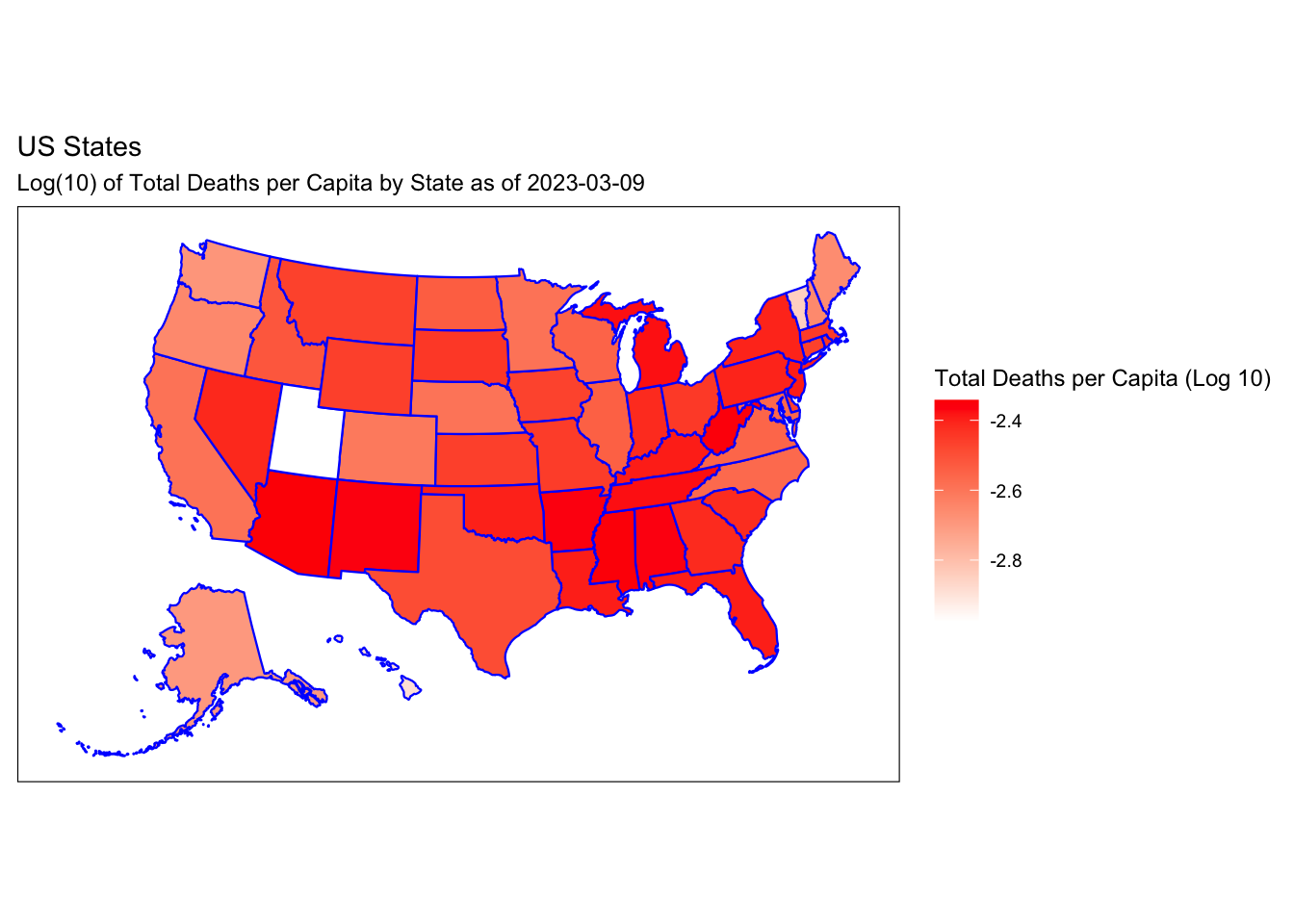
Rows: 72
Columns: 45
$ objectid <dbl> 4, 26, 33, 16, 25, 34, 72, 10, 63, 2, 19, 65, 46, 39, 42, 5…
$ gnocdc_lab <chr> "AUDUBON", "FRERET", "IBERVILLE", "DILLARD", "FLORIDA DEV",…
$ lup_lab <chr> "AUDUBON - UNIVERSITY", "FRERET", "IBERVILLE HOUSING DEV", …
$ neigh_id <chr> "10A", "11B", "6C", "3D", "8C", "13B", "13A", "11A", "13F",…
$ cnt_lup_la <dbl> 17, 3, 2, 6, 1, 5, 6, 7, 5, 3, 4, 3, 4, 2, 8, 3, 11, 7, 4, …
$ sum_pop100 <dbl> 17054, 1643, 1122, 5413, 160, 3104, 4742, 6039, 2295, 2289,…
$ sum_hu100 <dbl> 5862, 931, 647, 2483, 52, 1922, 2740, 3056, 1269, 1414, 153…
$ sum_totpop <dbl> 17054, 1643, 1122, 5413, 160, 3104, 4742, 6039, 2295, 2289,…
$ sum_white <dbl> 13466, 793, 87, 447, 2, 2069, 3470, 2491, 624, 1664, 1633, …
$ sum_black <dbl> 434, 627, 967, 4593, 154, 620, 682, 2735, 1457, 348, 618, 3…
$ sum_amer_i <dbl> 70, 14, 7, 16, 0, 9, 11, 33, 4, 13, 8, 9, 3, 3, 21, 16, 12,…
$ sum_asian <dbl> 500, 34, 8, 28, 0, 51, 109, 95, 38, 18, 45, 64, 17, 40, 250…
$ sum_pac_is <dbl> 29, 0, 0, 2, 1, 0, 4, 0, 1, 4, 2, 1, 0, 0, 0, 2, 6, 0, 2, 0…
$ sum_other_ <dbl> 322, 47, 10, 94, 0, 84, 83, 242, 20, 29, 63, 59, 37, 32, 17…
$ sum_multi_ <dbl> 2233, 128, 43, 233, 3, 271, 383, 443, 151, 213, 232, 225, 1…
$ sum_totp_2 <dbl> 17054, 1643, 1122, 5413, 160, 3104, 4742, 6039, 2295, 2289,…
$ sum_hisp_l <dbl> 3057, 162, 34, 174, 2, 274, 367, 631, 110, 165, 209, 262, 9…
$ sum_non_hi <dbl> 13997, 1481, 1088, 5239, 158, 2830, 4375, 5408, 2185, 2124,…
$ sum_totp_3 <dbl> 15066, 1408, 816, 4582, 107, 2716, 3927, 4889, 1769, 1897, …
$ sum_white_ <dbl> 11894, 695, 73, 385, 2, 1821, 2888, 2094, 571, 1422, 1423, …
$ sum_black_ <dbl> 384, 534, 688, 3912, 102, 547, 585, 2198, 1027, 272, 522, 3…
$ sum_amerin <dbl> 63, 12, 7, 13, 0, 9, 10, 22, 3, 13, 0, 4, 2, 2, 18, 11, 4, …
$ sum_asian_ <dbl> 427, 30, 6, 28, 0, 46, 85, 89, 38, 17, 32, 59, 16, 33, 241,…
$ sum_pacisl <dbl> 29, 0, 0, 2, 1, 0, 4, 0, 1, 2, 2, 1, 0, 0, 0, 2, 6, 0, 2, 0…
$ sum_othe_2 <dbl> 298, 34, 9, 74, 0, 67, 78, 165, 17, 25, 51, 45, 25, 18, 162…
$ sum_mult_2 <dbl> 1971, 103, 33, 168, 2, 226, 277, 321, 112, 146, 165, 186, 6…
$ sum_totp_4 <dbl> 15066, 1408, 816, 4582, 107, 2716, 3927, 4889, 1769, 1897, …
$ sum_hisp_2 <dbl> 2877, 117, 31, 112, 1, 226, 299, 442, 84, 115, 148, 203, 56…
$ sum_non__2 <dbl> 12189, 1291, 785, 4470, 106, 2490, 3628, 4447, 1685, 1782, …
$ sum_total_ <dbl> 5862, 931, 647, 2483, 52, 1922, 2740, 3056, 1269, 1414, 153…
$ sum_occ_hu <dbl> 4923, 782, 604, 2175, 51, 1650, 2389, 2643, 1156, 1162, 132…
$ sum_vacant <dbl> 939, 149, 43, 308, 1, 272, 351, 413, 113, 252, 209, 322, 26…
$ sum_totp_5 <dbl> 5536, 30, 0, 709, 0, 0, 120, 9, 0, 7, 0, 365, 20, 0, 7, 7, …
$ sum_instgq <dbl> 162, 0, 0, 0, 0, 0, 108, 0, 0, 0, 0, 327, 0, 0, 0, 7, 0, 0,…
$ sum_adulco <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ sum_juvcor <dbl> 0, 0, 0, 0, 0, 0, 18, 0, 0, 0, 0, 0, 0, 0, 0, 7, 0, 0, 0, 0…
$ sum_nursho <dbl> 162, 0, 0, 0, 0, 0, 90, 0, 0, 0, 0, 295, 0, 0, 0, 0, 0, 0, …
$ sum_othins <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 32, 0, 0, 0, 0, 0, 0, 0, 0…
$ sum_nonins <dbl> 5374, 30, 0, 709, 0, 0, 12, 9, 0, 7, 0, 38, 20, 0, 7, 0, 10…
$ sum_col_do <dbl> 5344, 0, 0, 707, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, …
$ sum_milita <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ sum_othnon <dbl> 30, 30, 0, 2, 0, 0, 12, 9, 0, 7, 0, 38, 20, 0, 7, 0, 10, 5,…
$ shape_star <dbl> 63257823, 5464058, 1747063, 26475756, 1540363, 23457801, 38…
$ shape_stle <dbl> 39015.654, 9710.452, 5408.459, 20830.352, 5959.030, 26507.6…
$ geometry <POLYGON [°]> POLYGON ((-90.10964 29.9413..., POLYGON ((-90.10331…