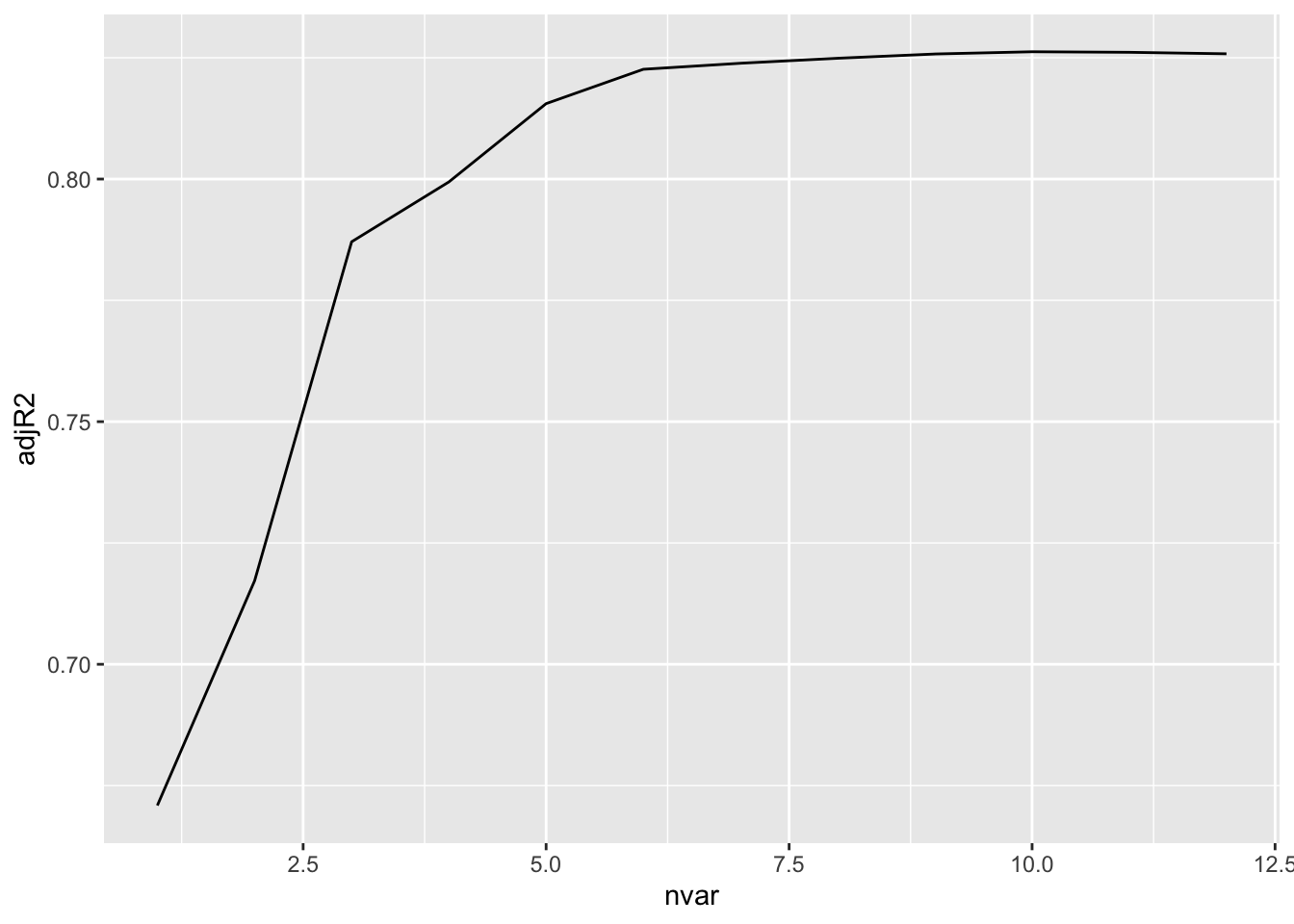
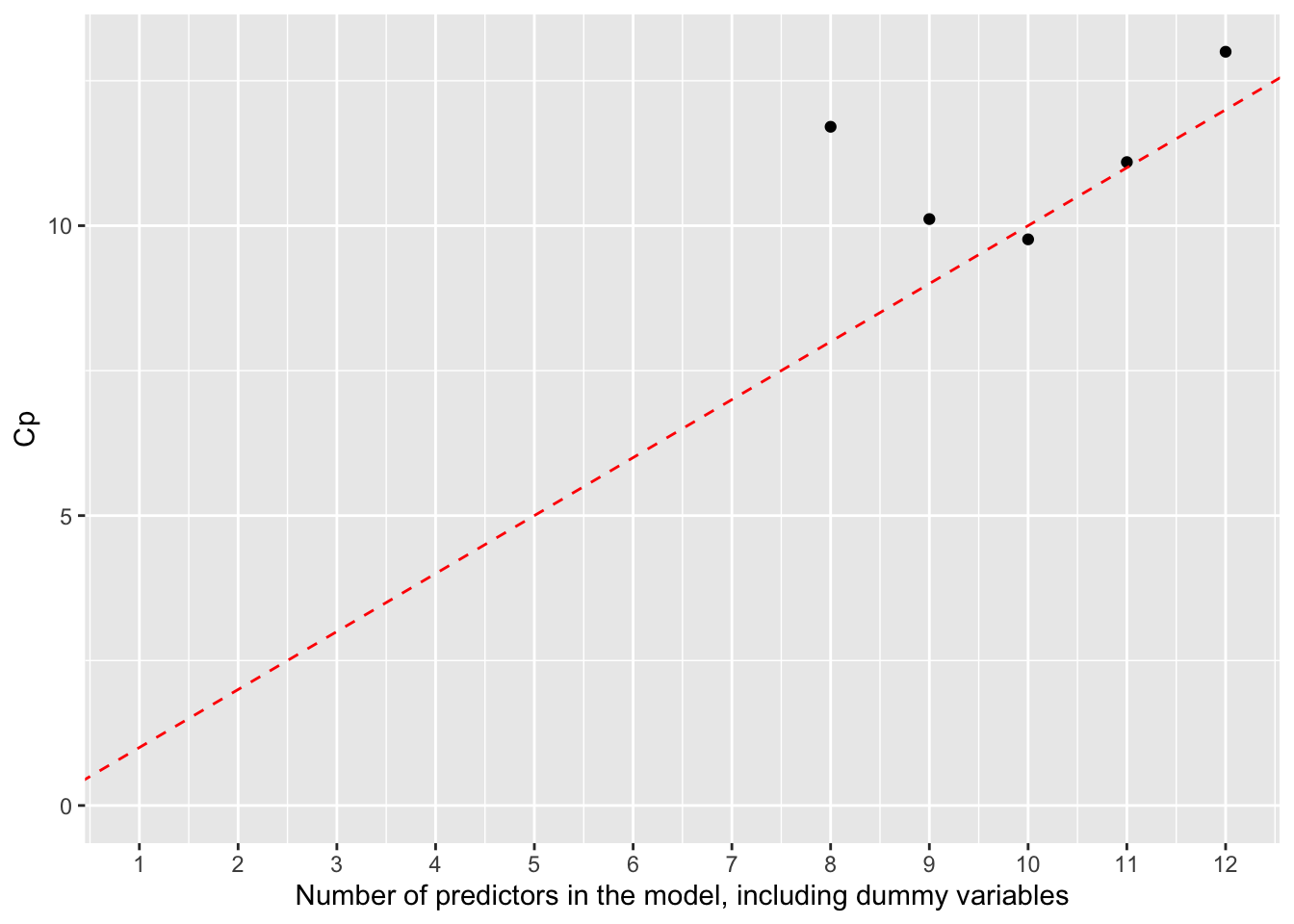
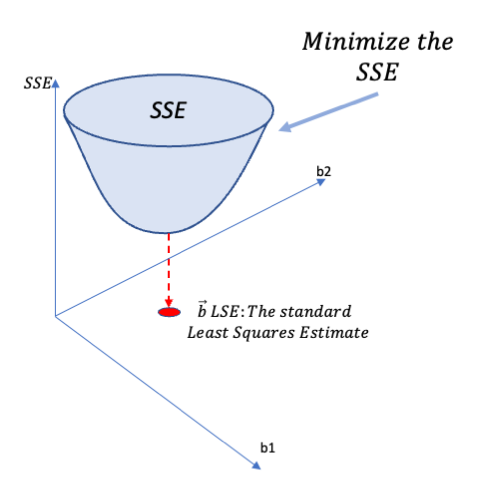
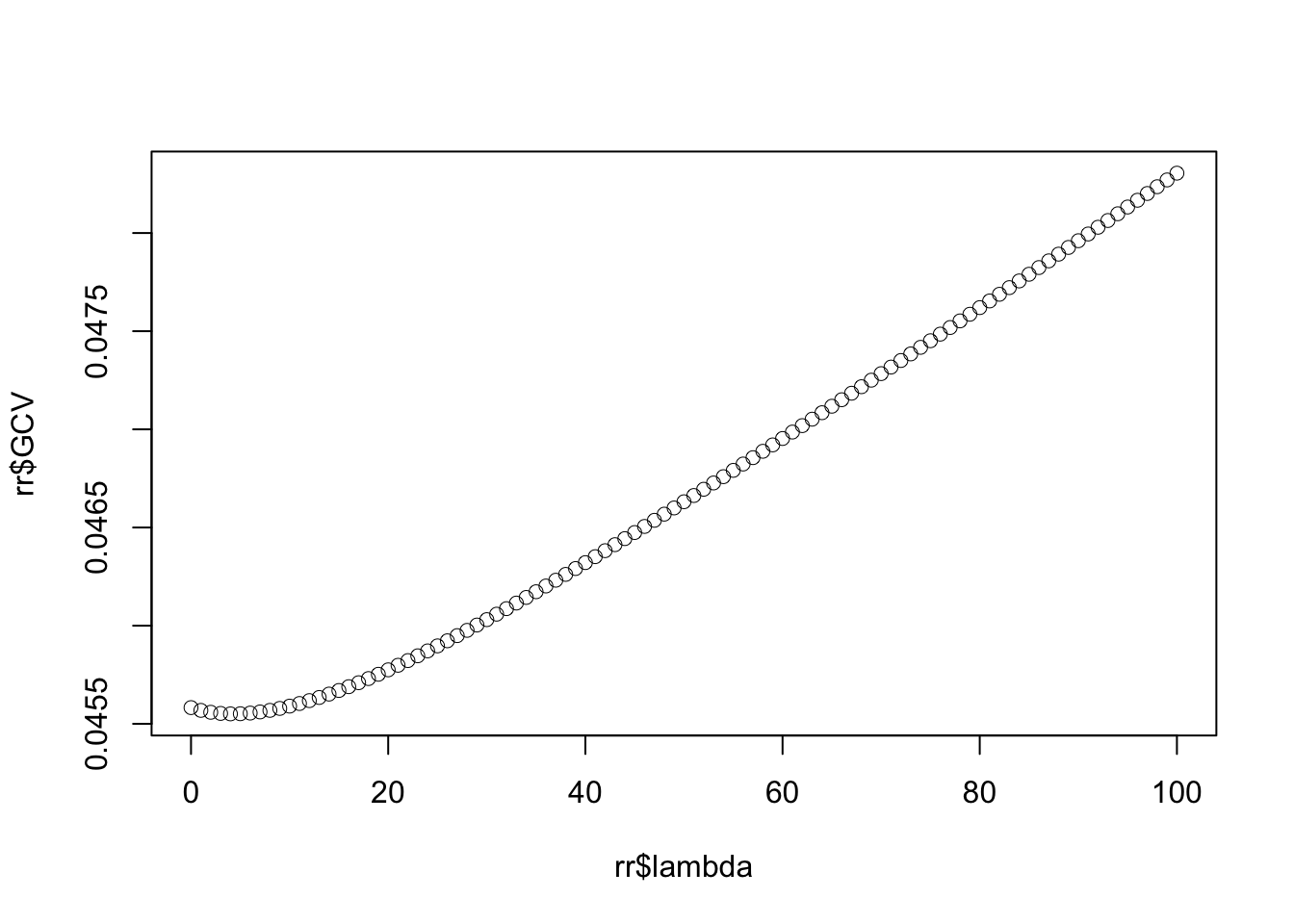
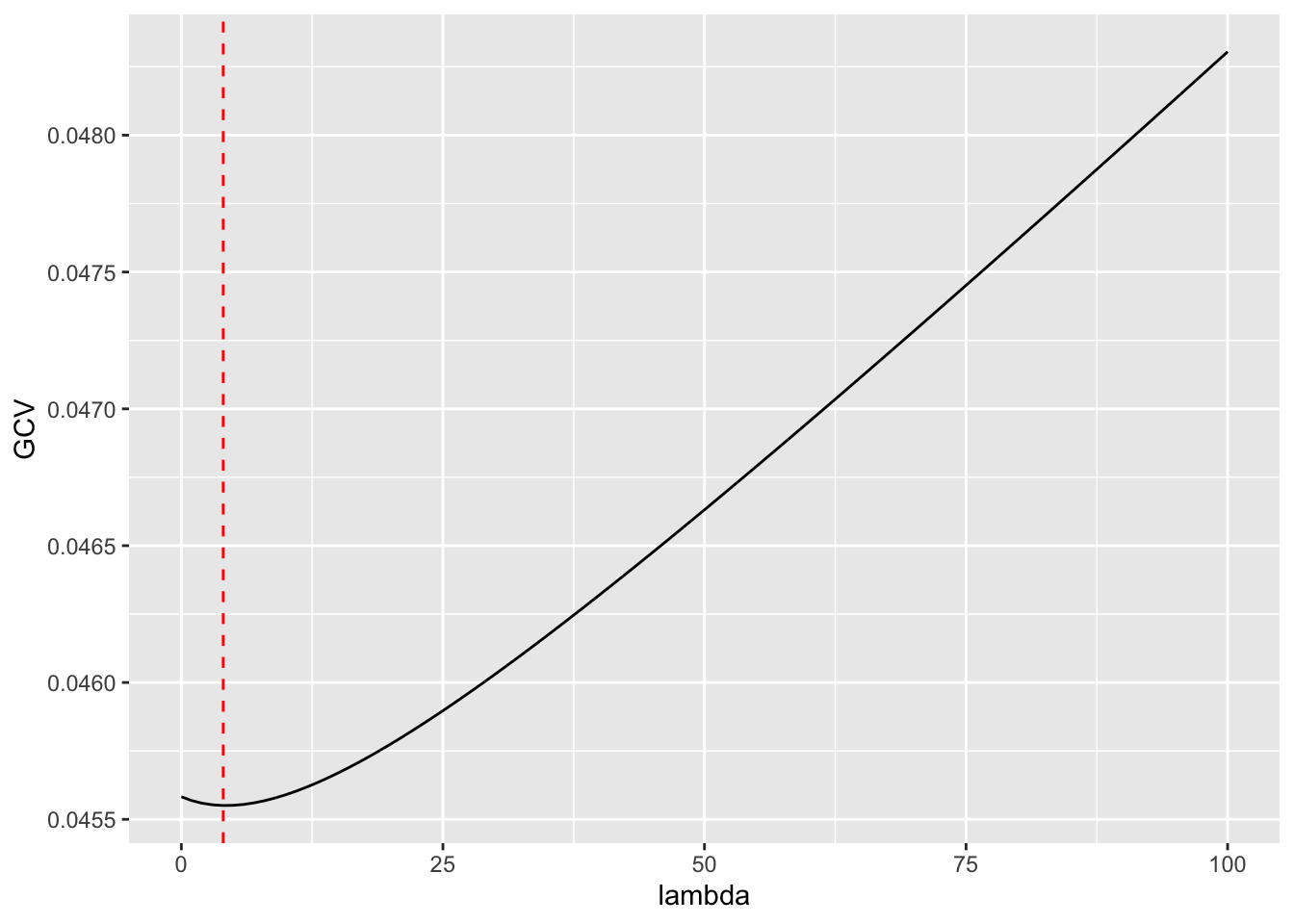
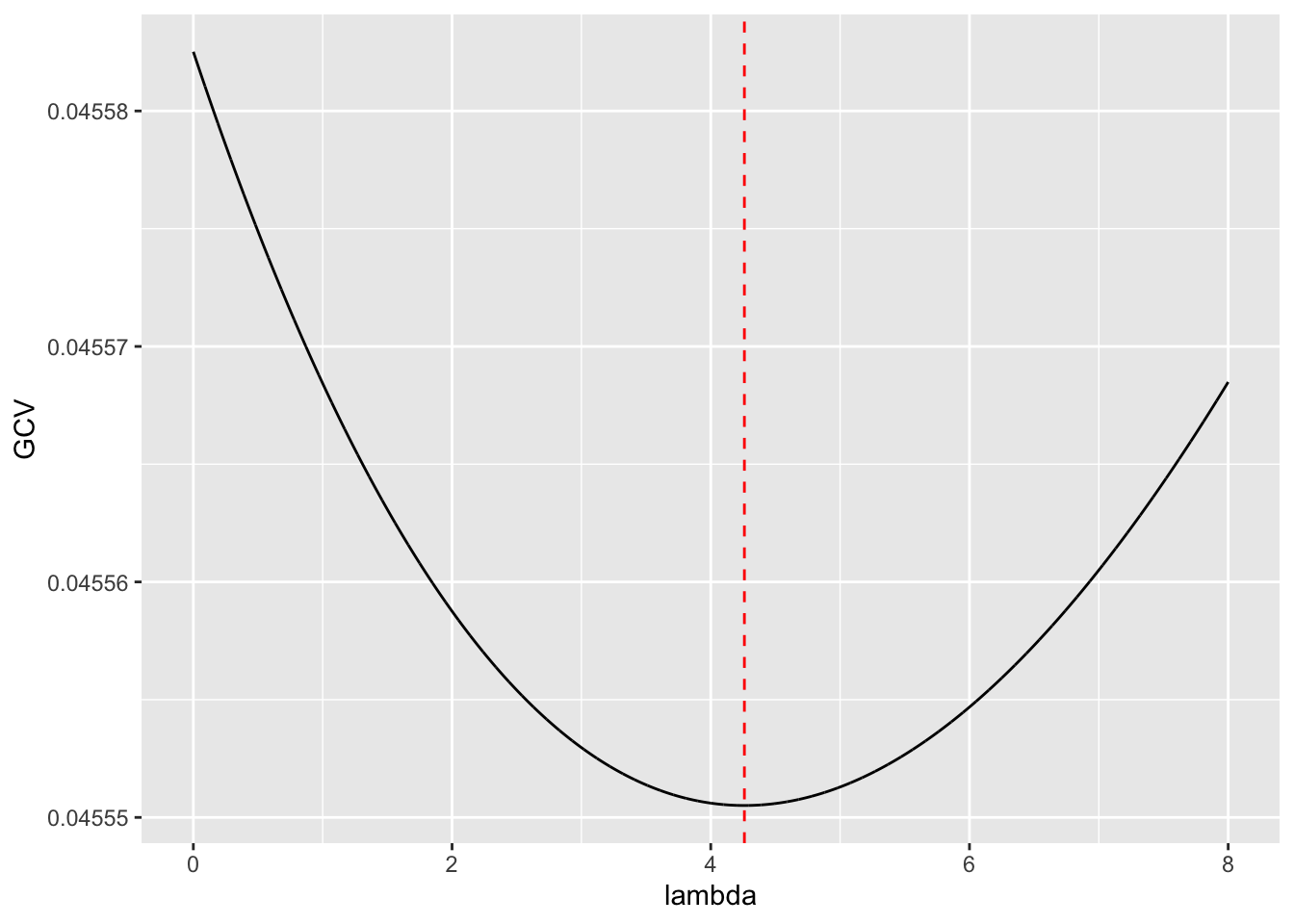
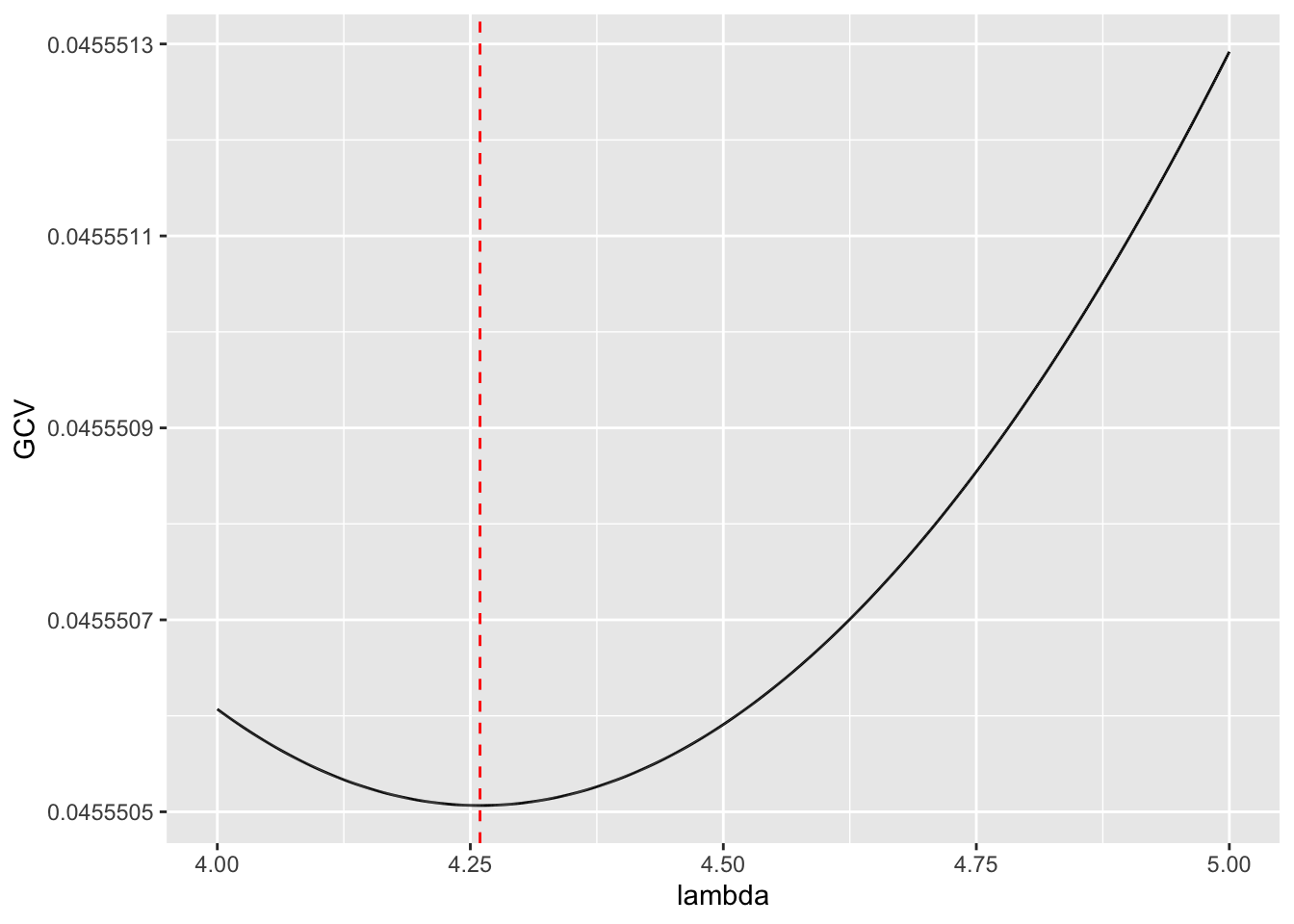
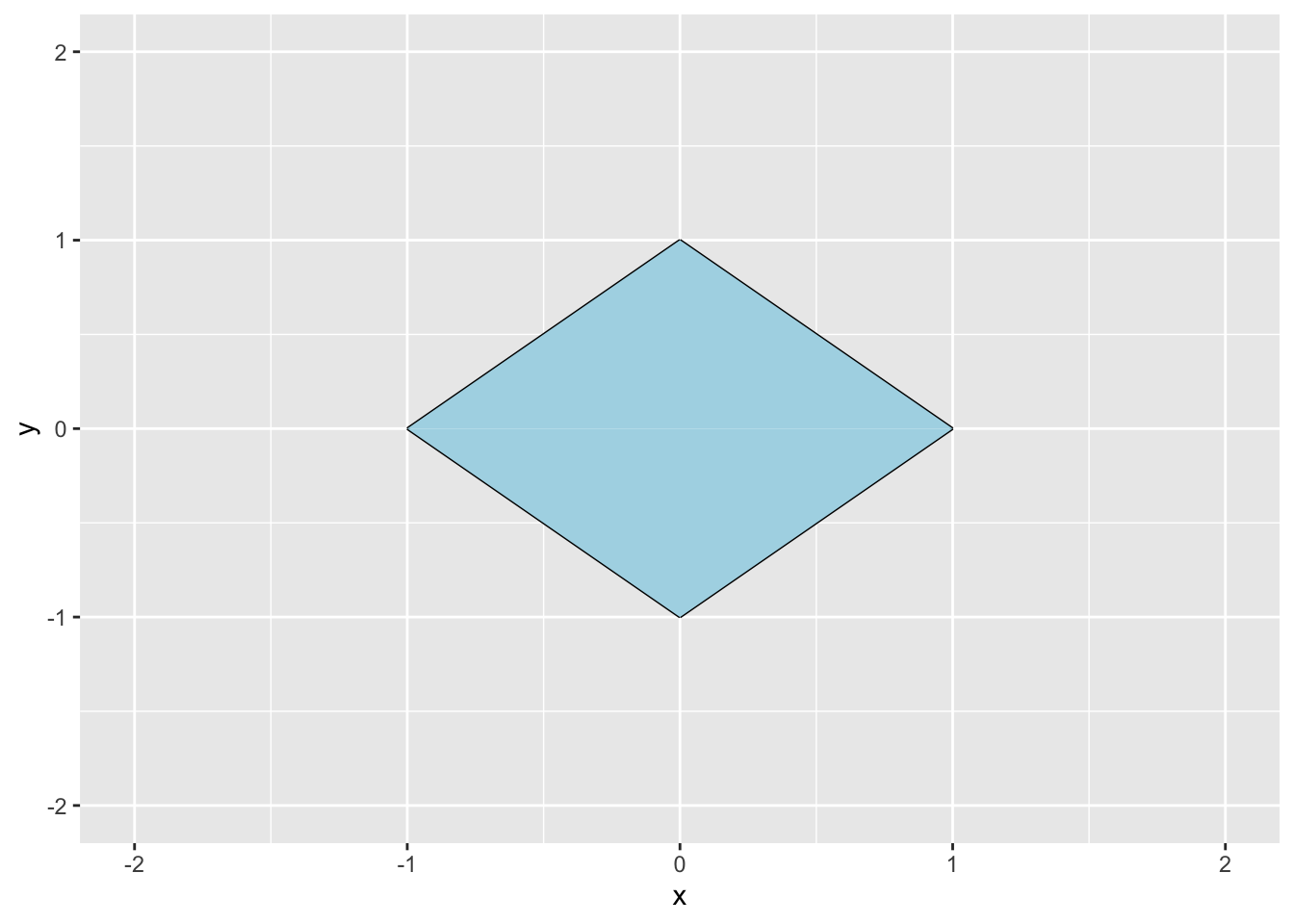
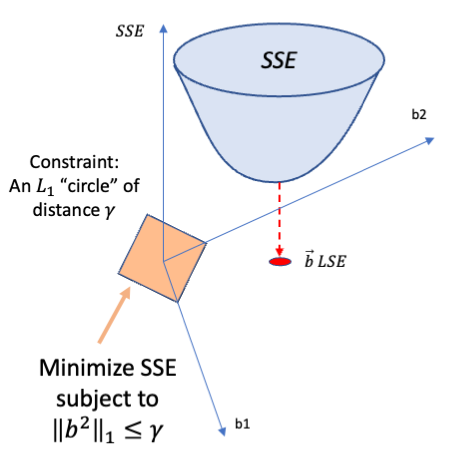
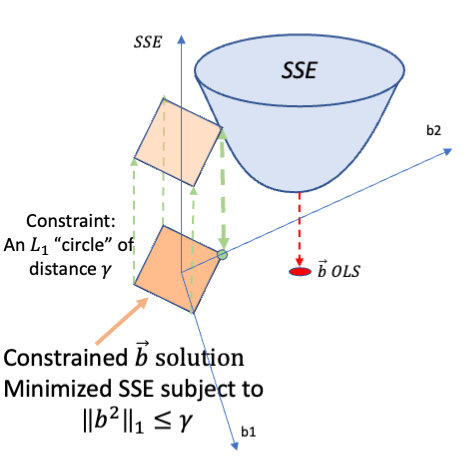
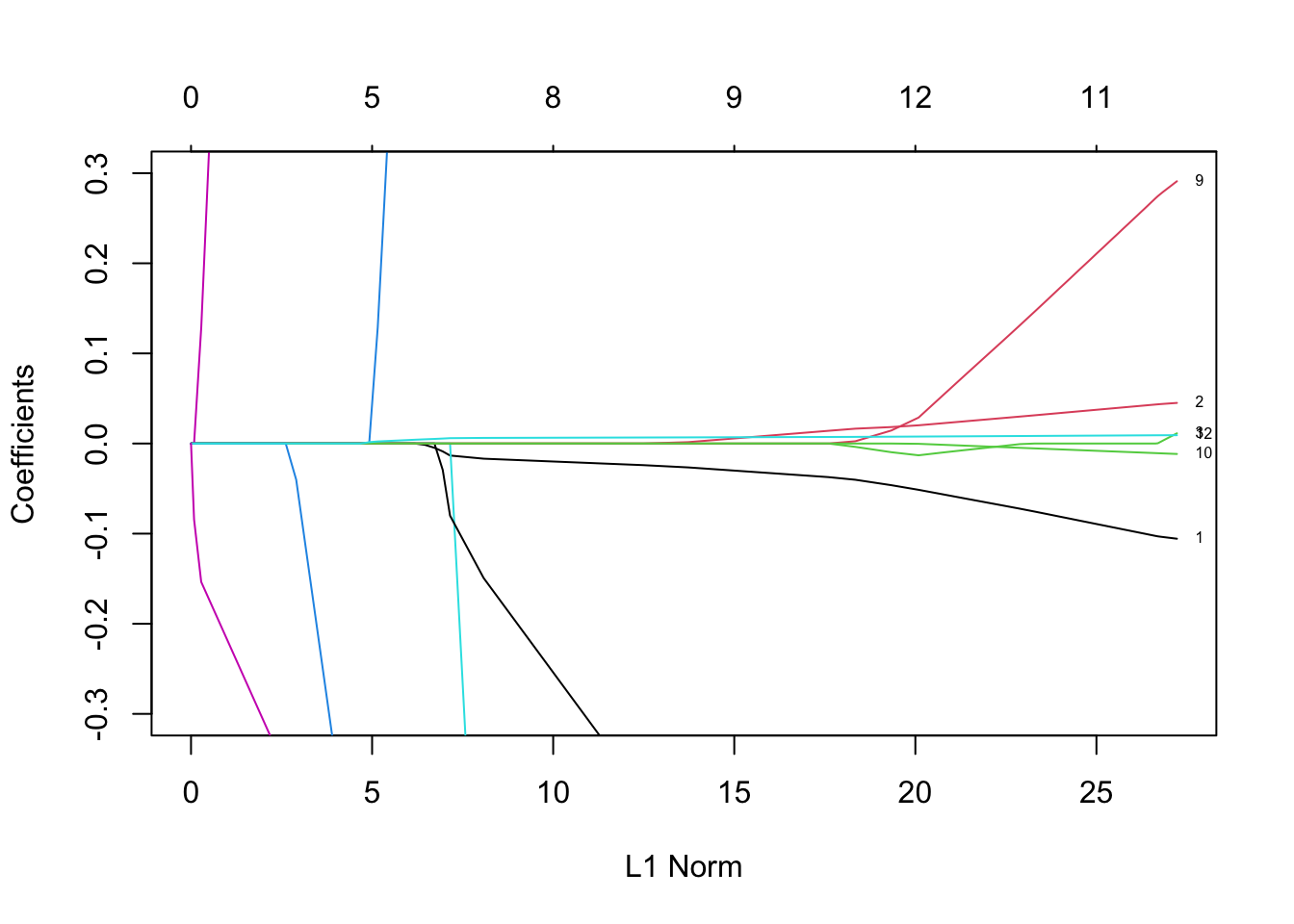
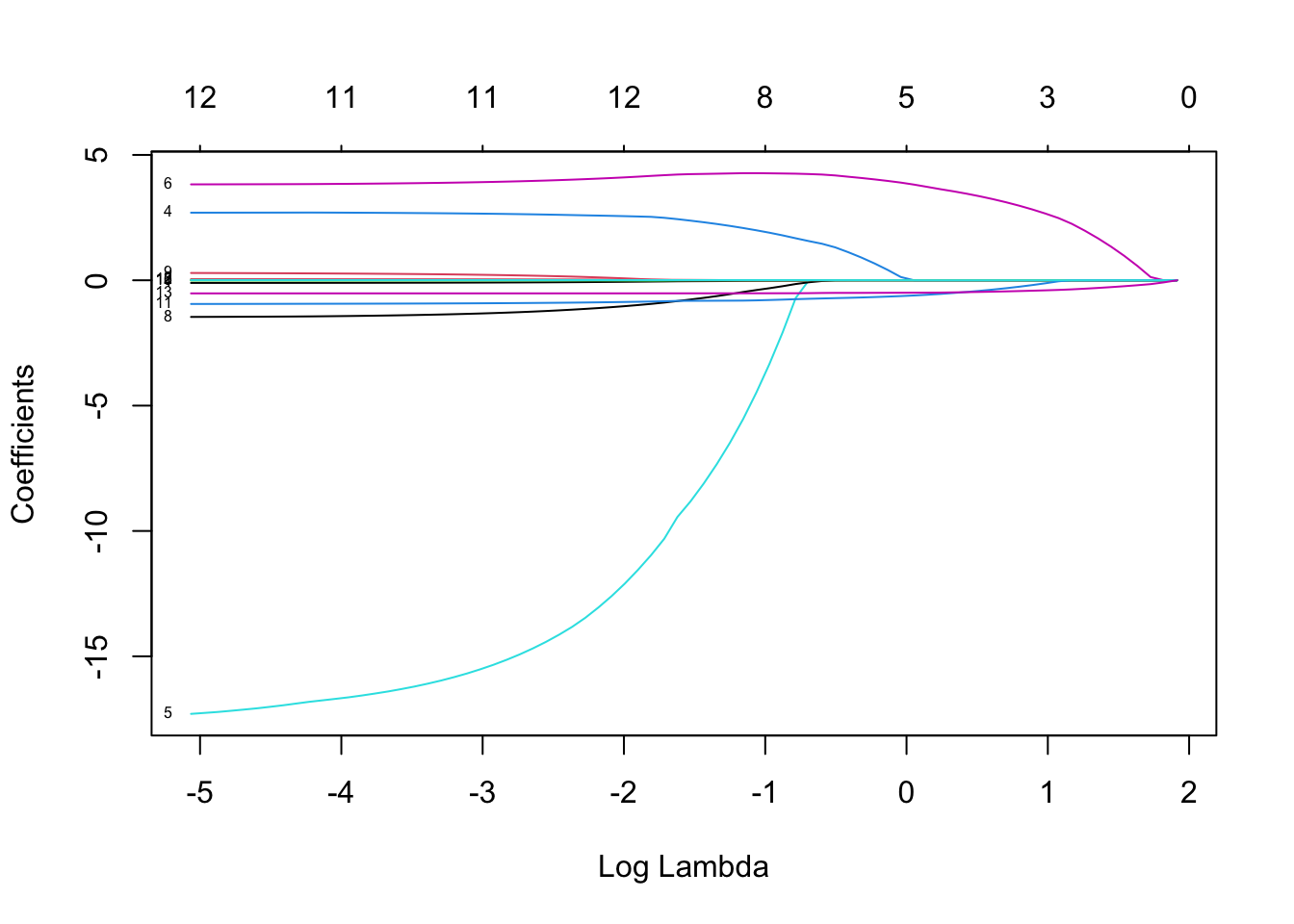
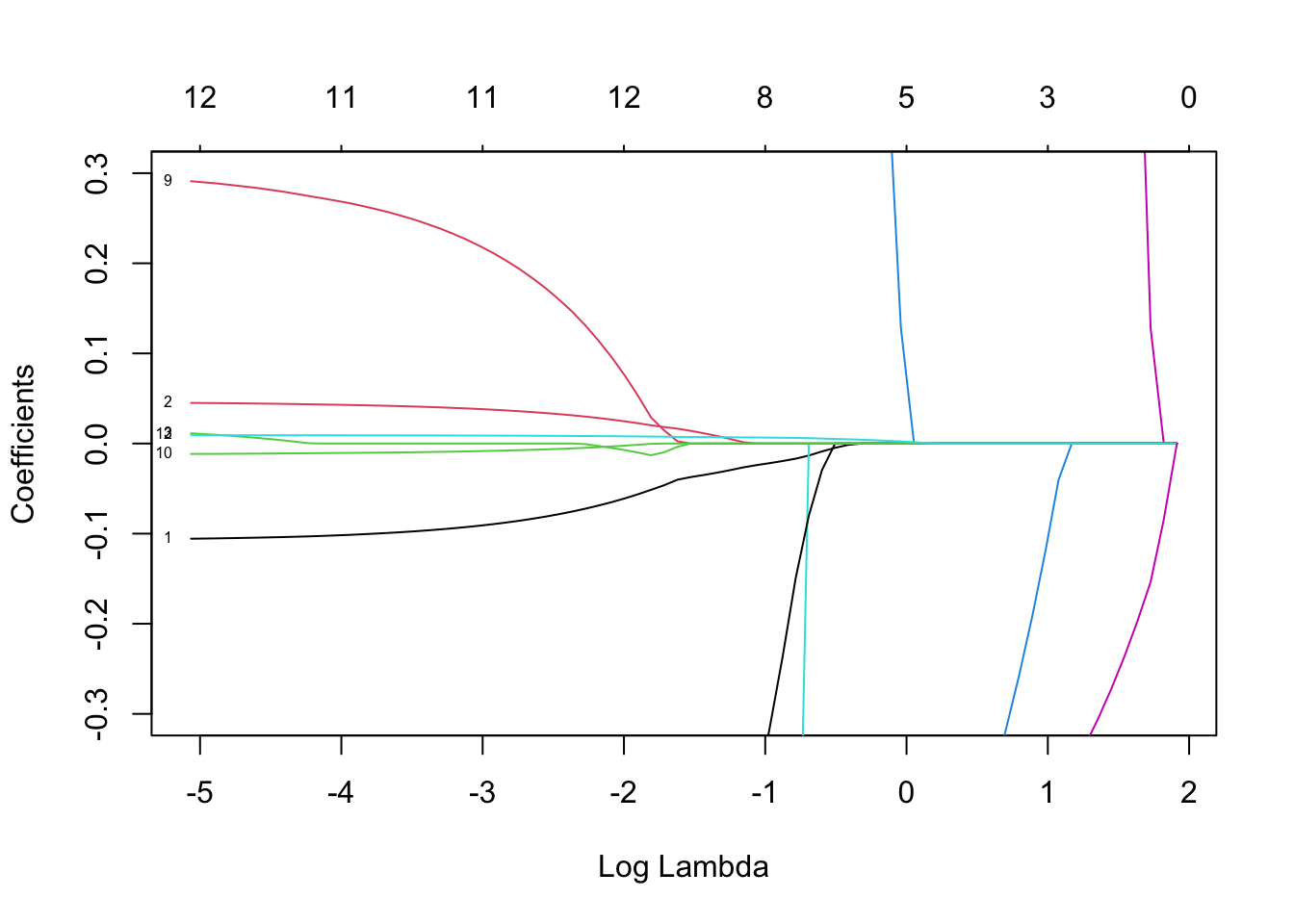
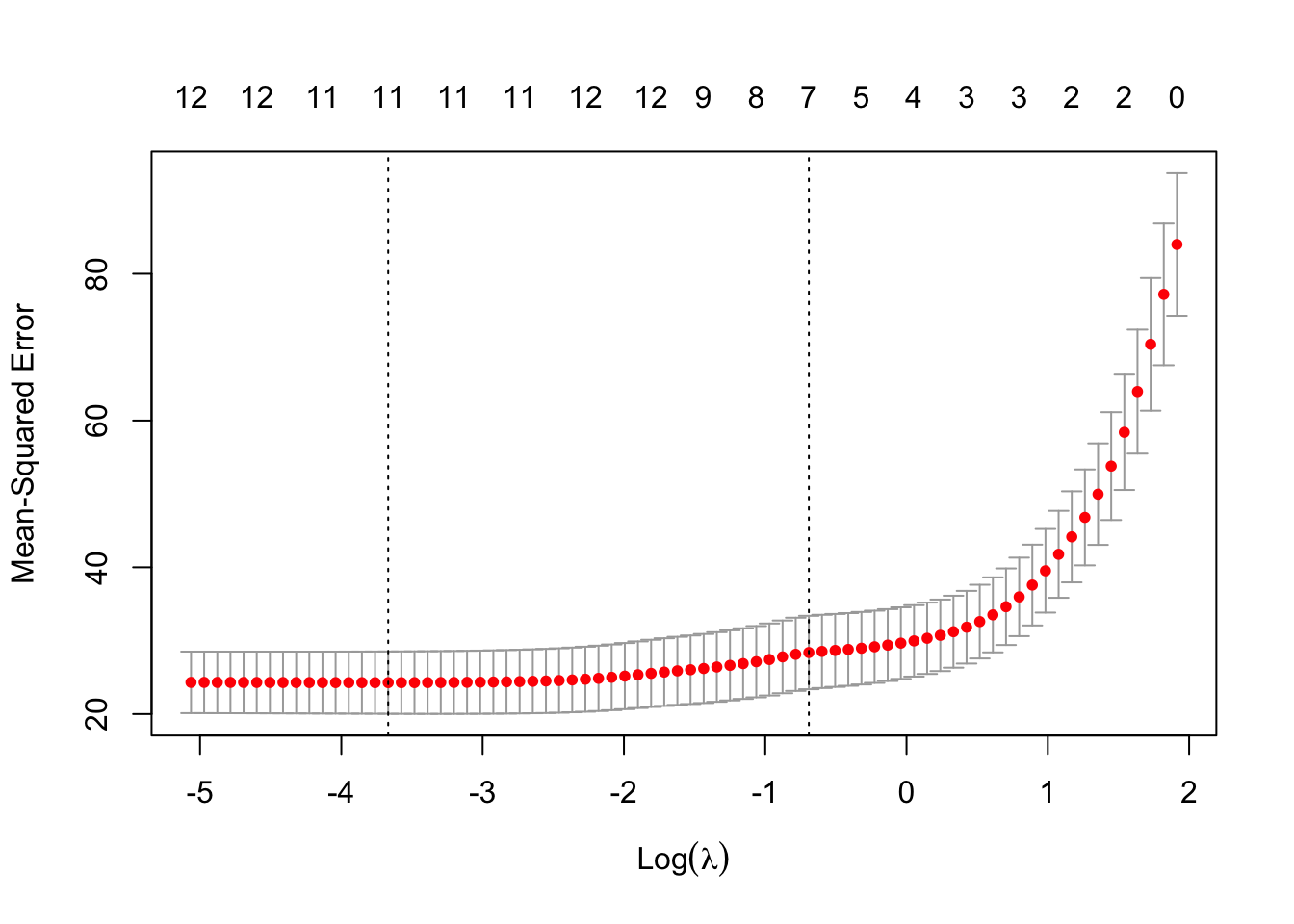
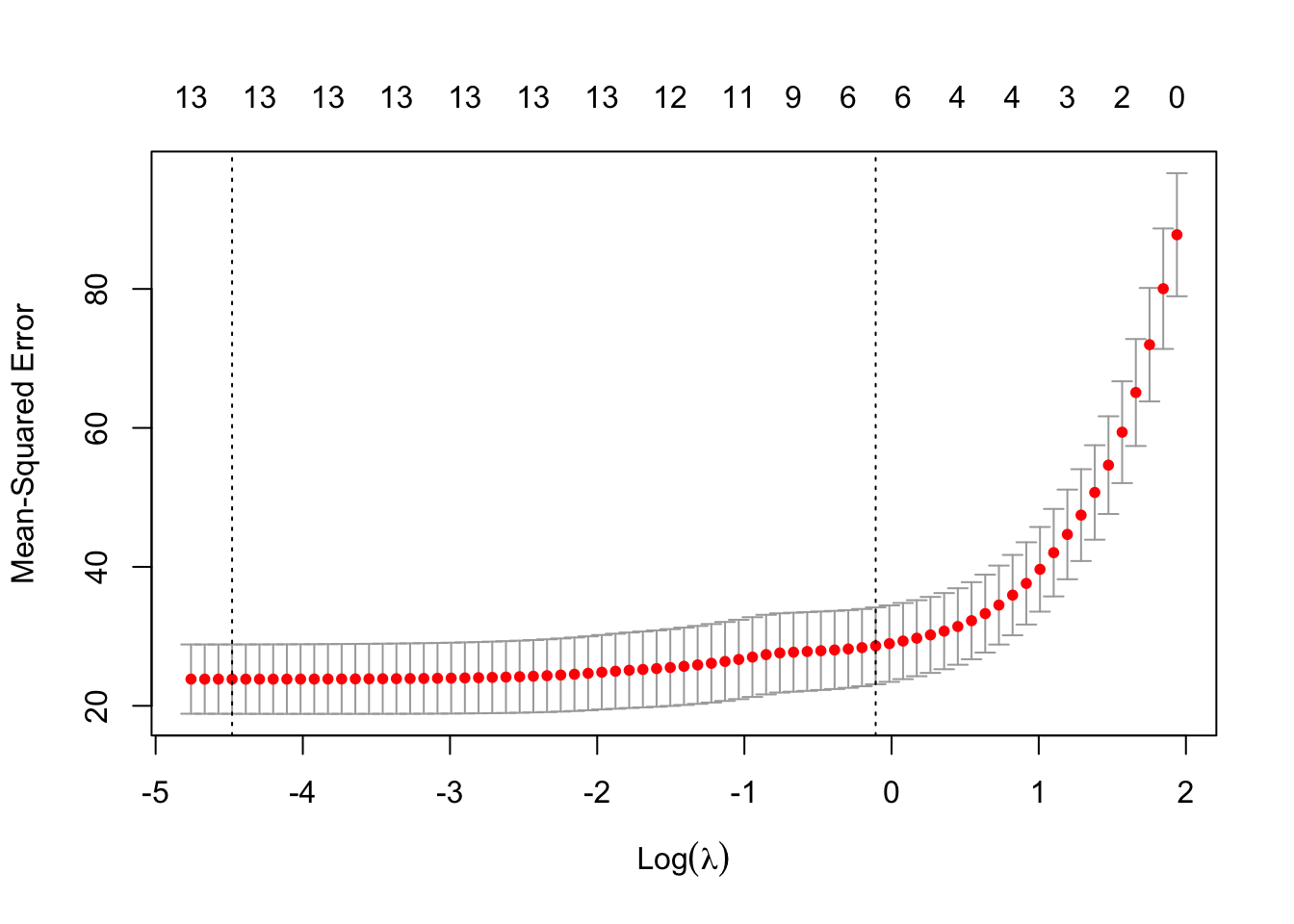
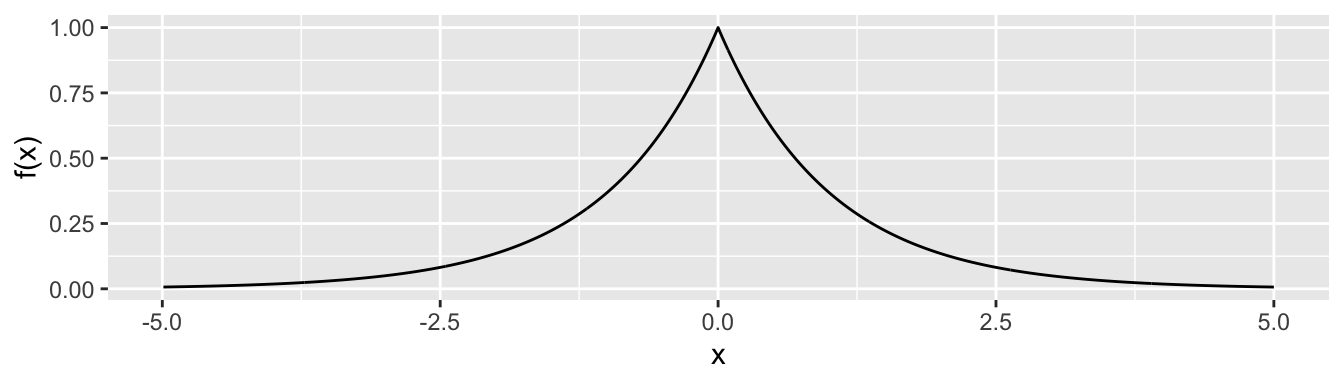
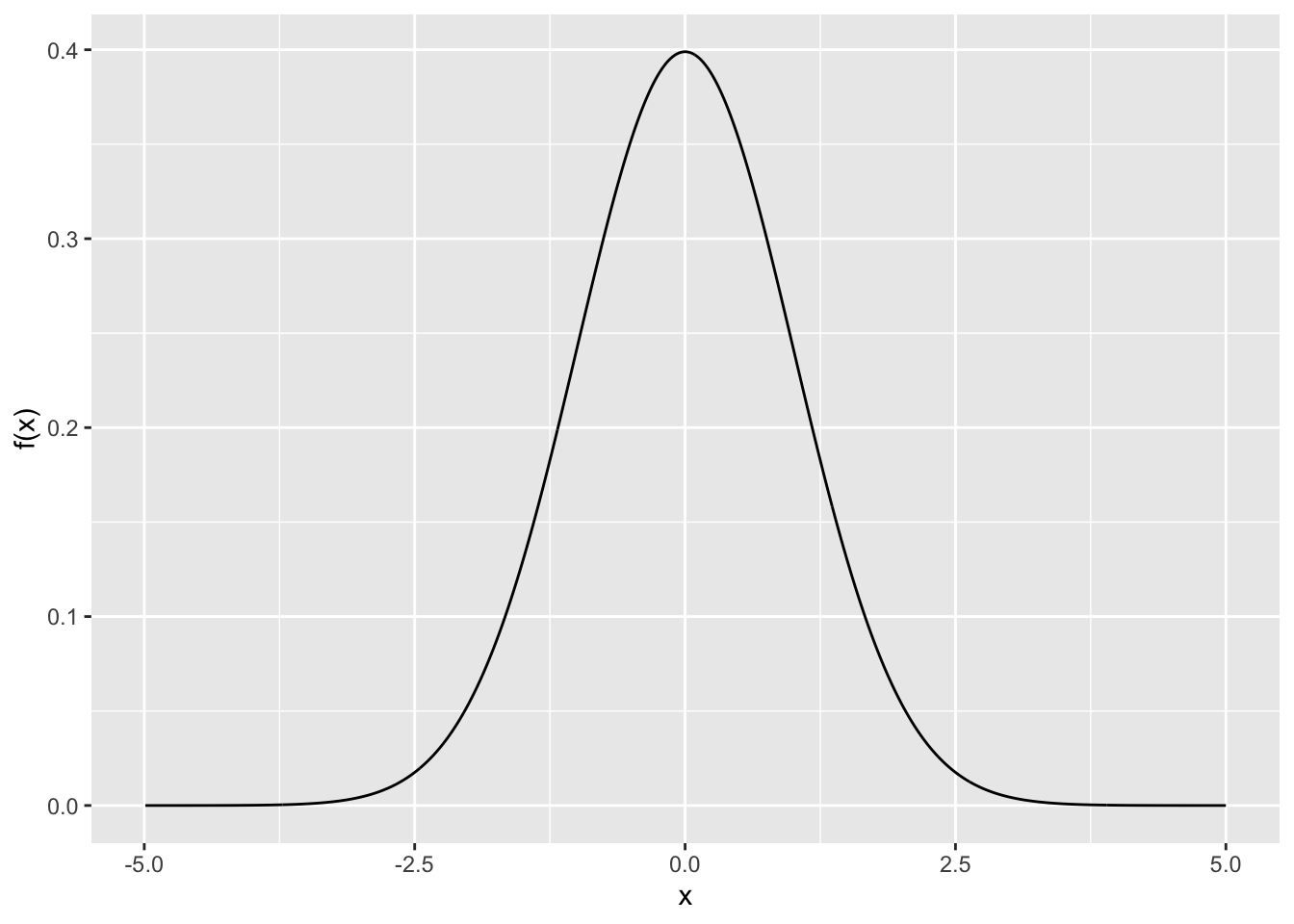
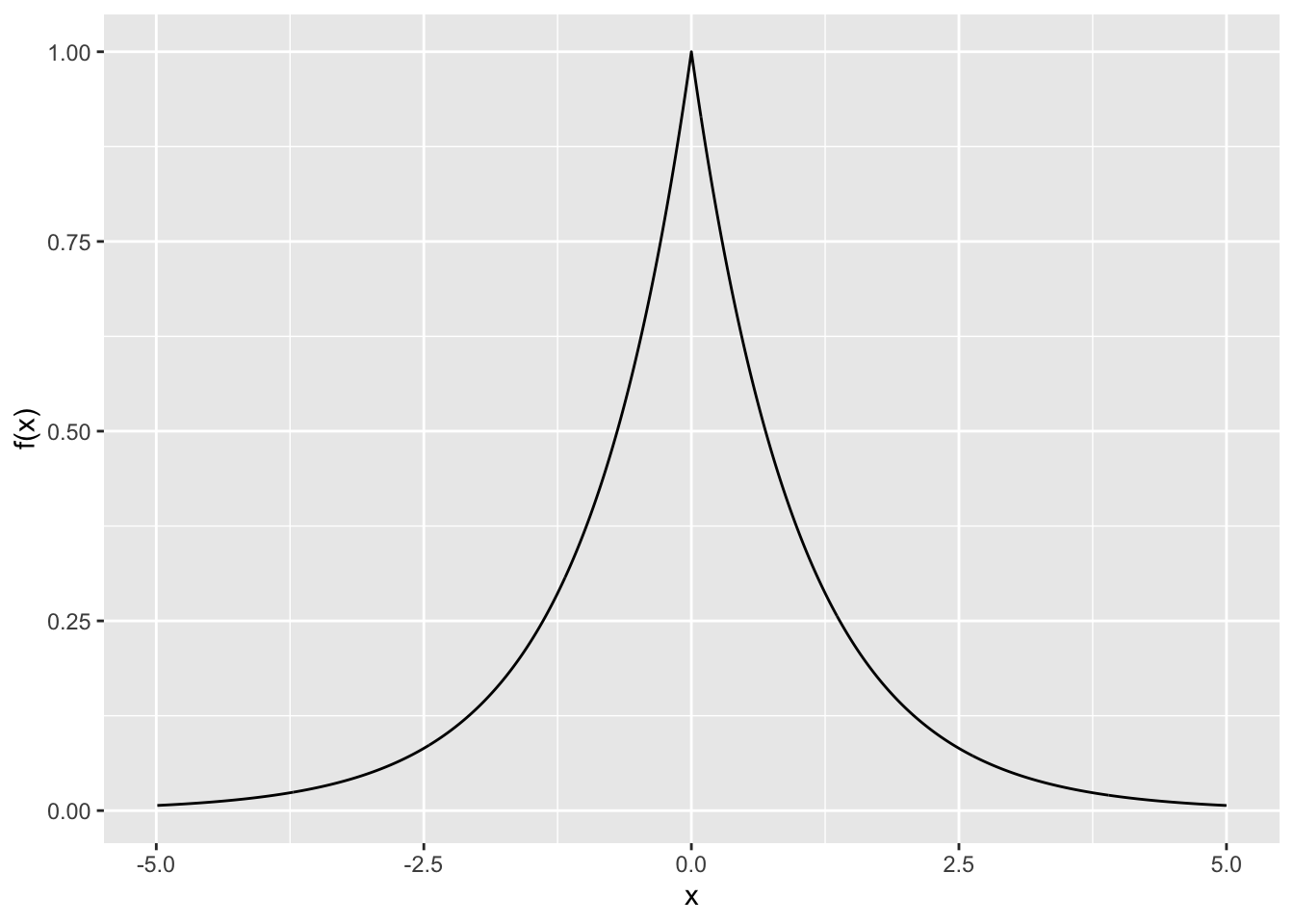
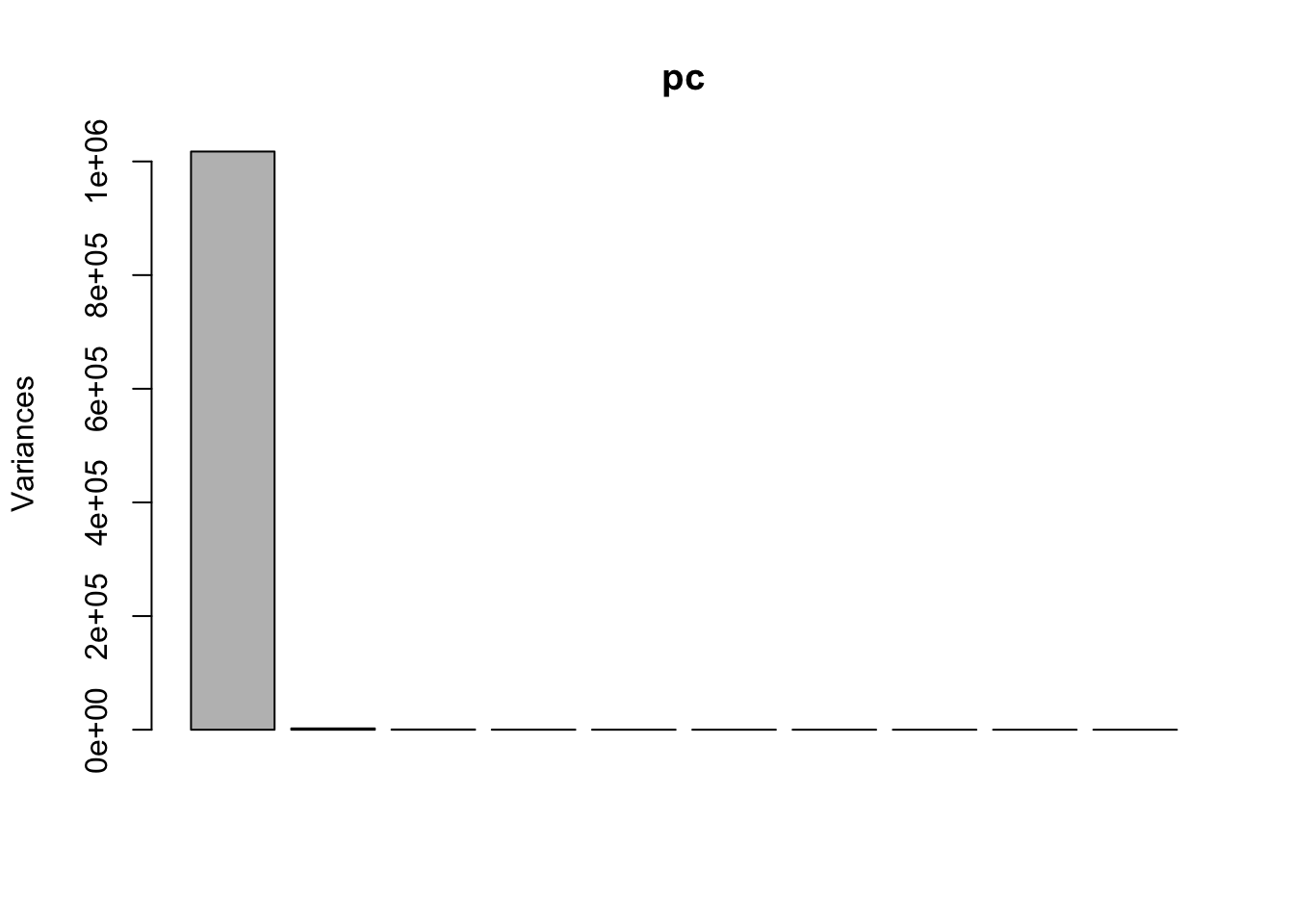
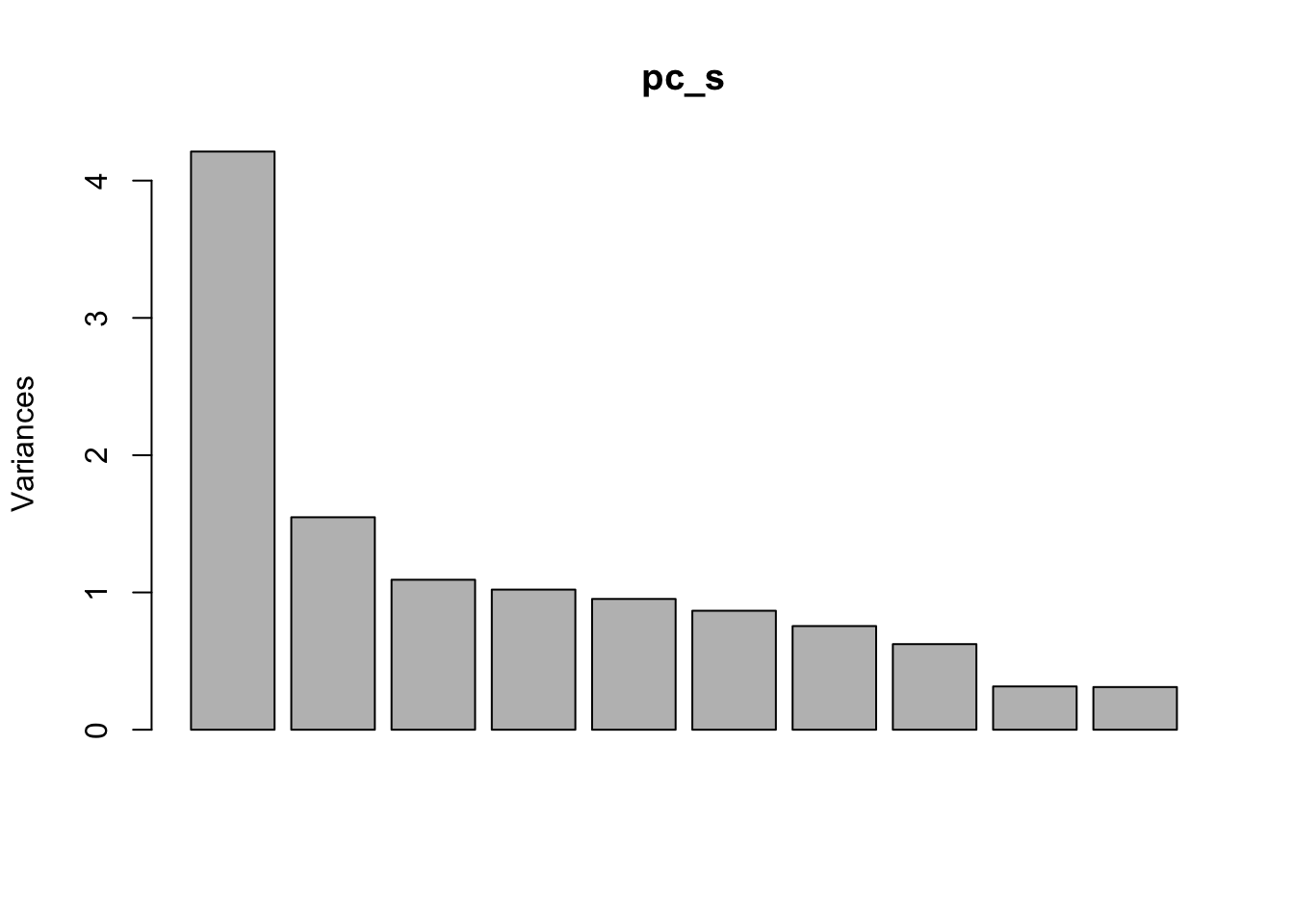
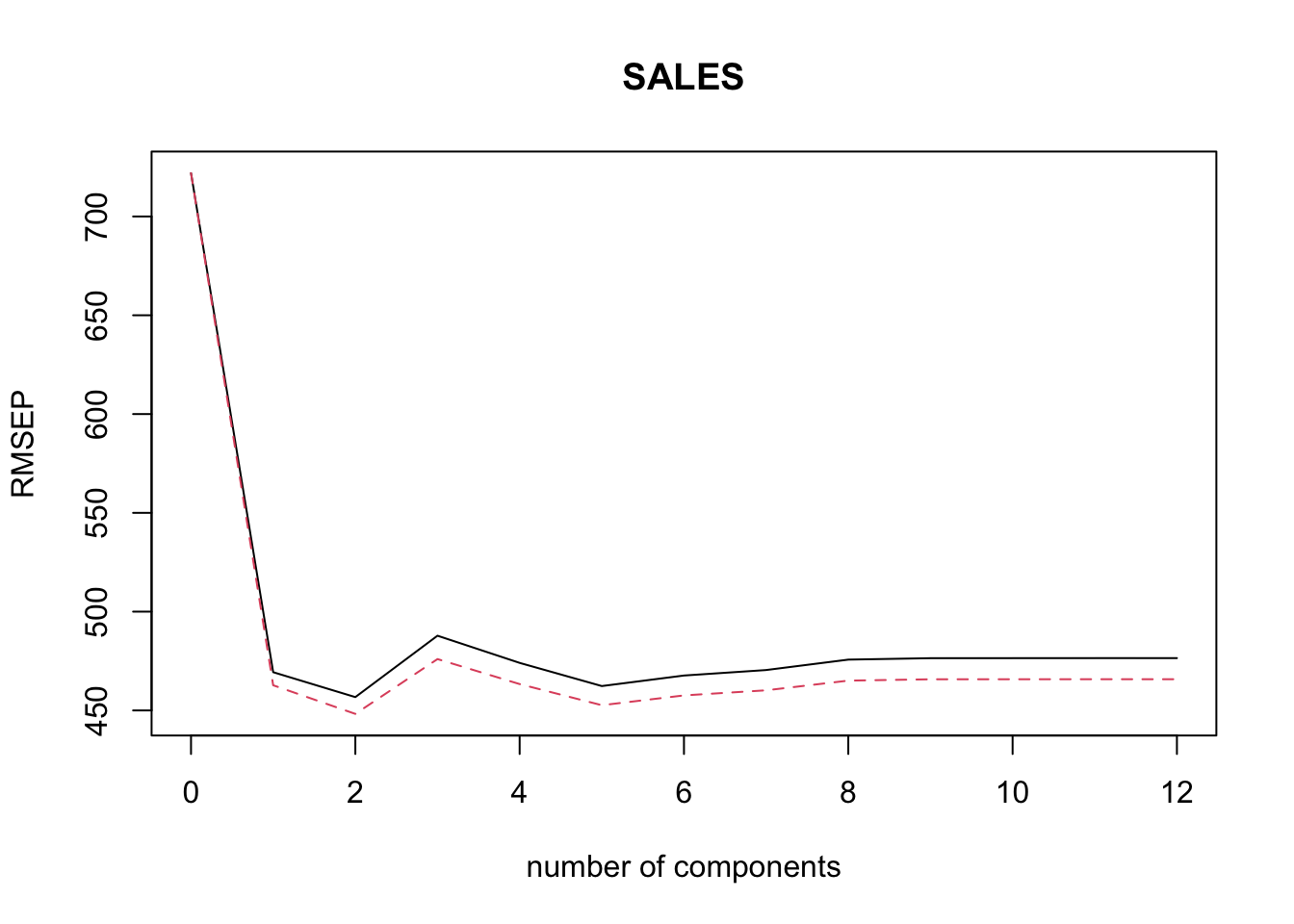
Rows: 279
Columns: 13
$ OUTLOOK <dbl> 2, 4, 5, 5, 2, 3, 2, 3, 4, 4, 4, 4, 4, 3, 3, 4, 3, 2, 5, 3, 3…
$ SALES <dbl> 480, 507, 210, 246, 148, 50, 72, 99, 160, 243, 200, 1000, 350…
$ NEWCAP <dbl> 0, 22, 25, NA, NA, NA, 0, 7, 5, 7, 3, 20, NA, 0, 10, 8, 0, NA…
$ VALUE <dbl> 600, 375, 275, 80, 85, 135, 125, 150, 85, 150, 225, 1500, NA,…
$ COSTGOOD <dbl> 35, 59, 40, 43, 45, 40, 85, 43, NA, 38, 42, 20, 31, 50, 50, N…
$ WAGES <dbl> 25, 20, 24, 30, 35, 30, 10, 25, NA, 15, 22, 20, 35, 26, 40, N…
$ ADS <dbl> 2, 5, 3, 1, 1, 10, 5, 1, NA, 2, 2, 10, NA, 2, 10, NA, 4, 5, 1…
$ TYPEFOOD <dbl> 2, 2, 1, 1, 3, 2, 2, 2, NA, 2, 1, 1, 1, 2, 2, NA, 1, 1, 2, 2,…
$ SEATS <dbl> 200, 150, 46, 28, 44, 50, 50, 130, NA, 50, 64, 240, 111, 125,…
$ OWNER <dbl> 3, 1, 1, 3, 1, 3, 1, 1, 2, 2, 1, 3, 3, 3, 1, 3, 3, 3, 3, 1, 2…
$ FTEMPL <dbl> 8, 6, 0, 2, NA, 2, 0, 1, 2, 2, 3, 30, 10, 6, 4, 13, 7, 20, 1,…
$ PTEMPL <dbl> 30, 25, 17, 13, NA, NA, 5, 8, 10, 19, 12, 40, 19, 16, 28, 47,…
$ SIZE <dbl> 3, 2, 1, 1, NA, NA, 1, 1, 1, 2, 1, 3, 2, 2, 2, 3, 2, 3, 1, 2,…