Show code
ISLRv2 Chapter 5 and Handouts
Cross-validation is a method for evaluating prediction performance in SML algorithms.
Recall, if we used training data to evaluate prediction performance, we would overestimate the true performance on new data due to inevitable over-fitting.
To evaluate prediction accuracy, we have separated our data in training and testing subsets.
This is a type of Resampling as we are drawing samples from the original data set.
There are different methods of Resampling. We have been using the validation set method.
We have used two of many possible Measures of Performance:
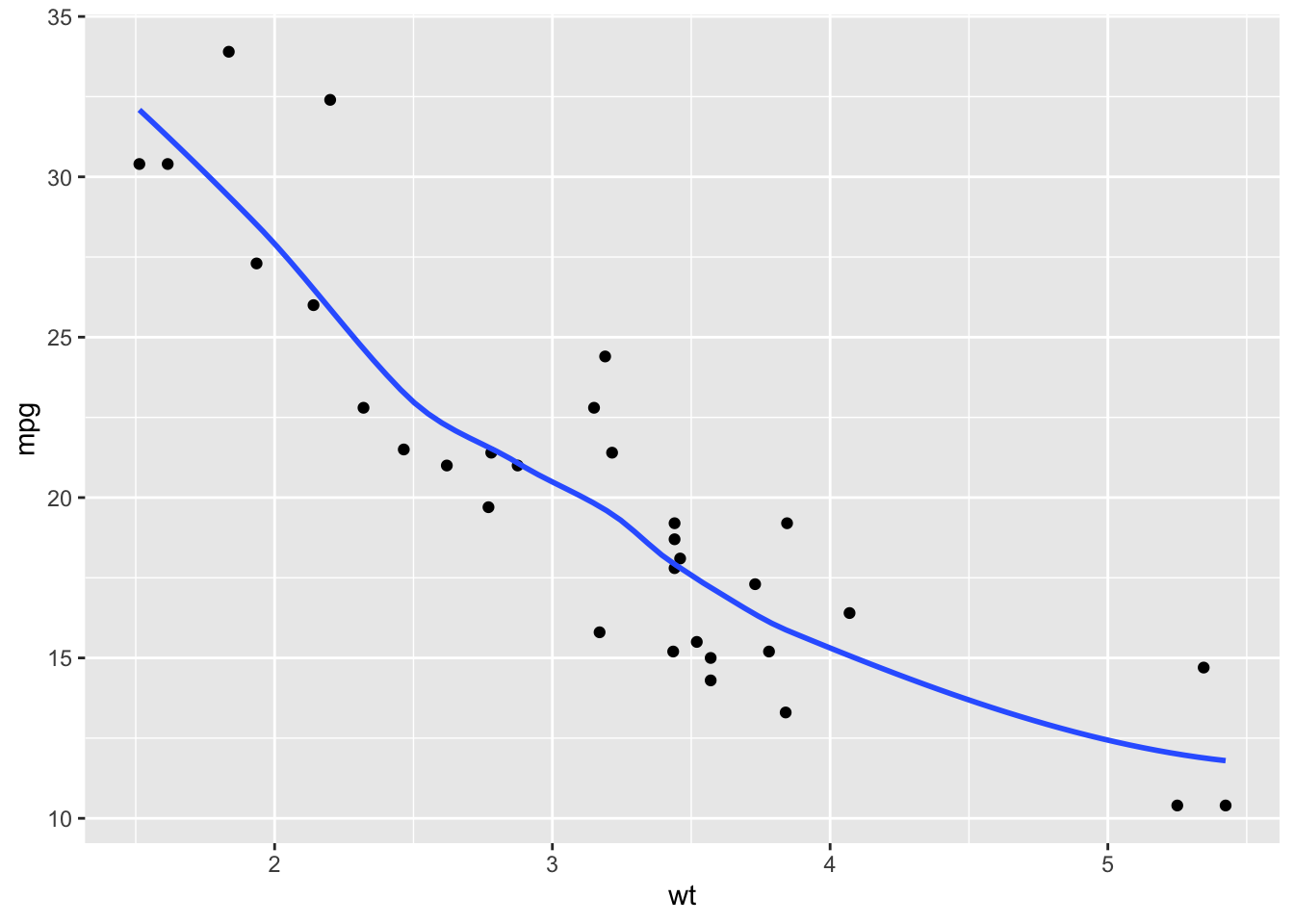
mtcars Data in RLet’s plot the data

We can see some non-linearity but perhaps that is just noise. Can we do better with a polynomial model?
We want to know what degree polynomial might be best for predicting the miles per gallon based on the weight of a new car.
We can use a for-loop to fit a series of polynomial regression models using the validation set method.
Recall, RMSE is the square root of the MSE which is \(\text{SSE}/n\).
It is more interpretable at times than MSE since it is in the un-squared units of the original \(Y\), e.g., mpg instead of mpg2.
set.seed(1234)
n <- nrow(mtcars)
Z <- sample(n, floor(n/2)) # setting up our re-sample
rmse <- vector(mode = "double", length = 10)
for (k in seq_along(rmse)) {
reg <- lm(mpg ~ poly(wt, k), data = mtcars, subset = Z) # training data
Yhat <- predict(reg, newdata = mtcars[-Z,]) # test data
Ytest = mtcars$mpg[-Z] # test data
rmse[[k]] <- sqrt(mean(Yhat - Ytest)^2)
}Let’s plot the RMSE versus \(k\) the degree of the polynomial.
Let’s look at the fitted values for miles per gallon. What do you notice?
Mazda RX4 Datsun 710 Merc 240D Merc 280
33.906610 168.235814 21.948461 17.313192
Merc 280C Merc 450SL Merc 450SLC Chrysler Imperial
17.313192 6.677194 3.065900 -490.900328
Fiat 128 Honda Civic Toyota Corolla Toyota Corona
232.388456 -821.758470 -277.777982 84.806589
Camaro Z28 Pontiac Firebird Porsche 914-2 Ford Pantera L
-1.181190 -1.495726 244.371357 22.325850 Let’s plot the fitted values against the training data and then the test data.
df <- tibble(wt_train = mtcars$wt[Z],
mpg_train = mtcars$mpg[Z],
Yhat_10 = Yhat)
ggplot(df, aes(wt_train, Yhat_10)) +
geom_point(color = "purple") +
geom_smooth(se = FALSE) +
geom_point(mapping = aes(wt_train, mpg_train), color = "green") +
ggtitle("Fitted values from the 10 degreee Polynomial on 16 Training Points")
df <- tibble(wt_test = mtcars$wt[-Z],
mpg_test = mtcars$mpg[-Z],
Yhat_10 = Yhat)
ggplot(df, aes(wt_test, Yhat_10)) +
geom_point(color = "red") +
geom_smooth(se = FALSE) +
geom_point(aes(wt_test, mpg_test)) +
ggtitle("Predicted values from the 10 degreee Polynomial on 16 Test Points")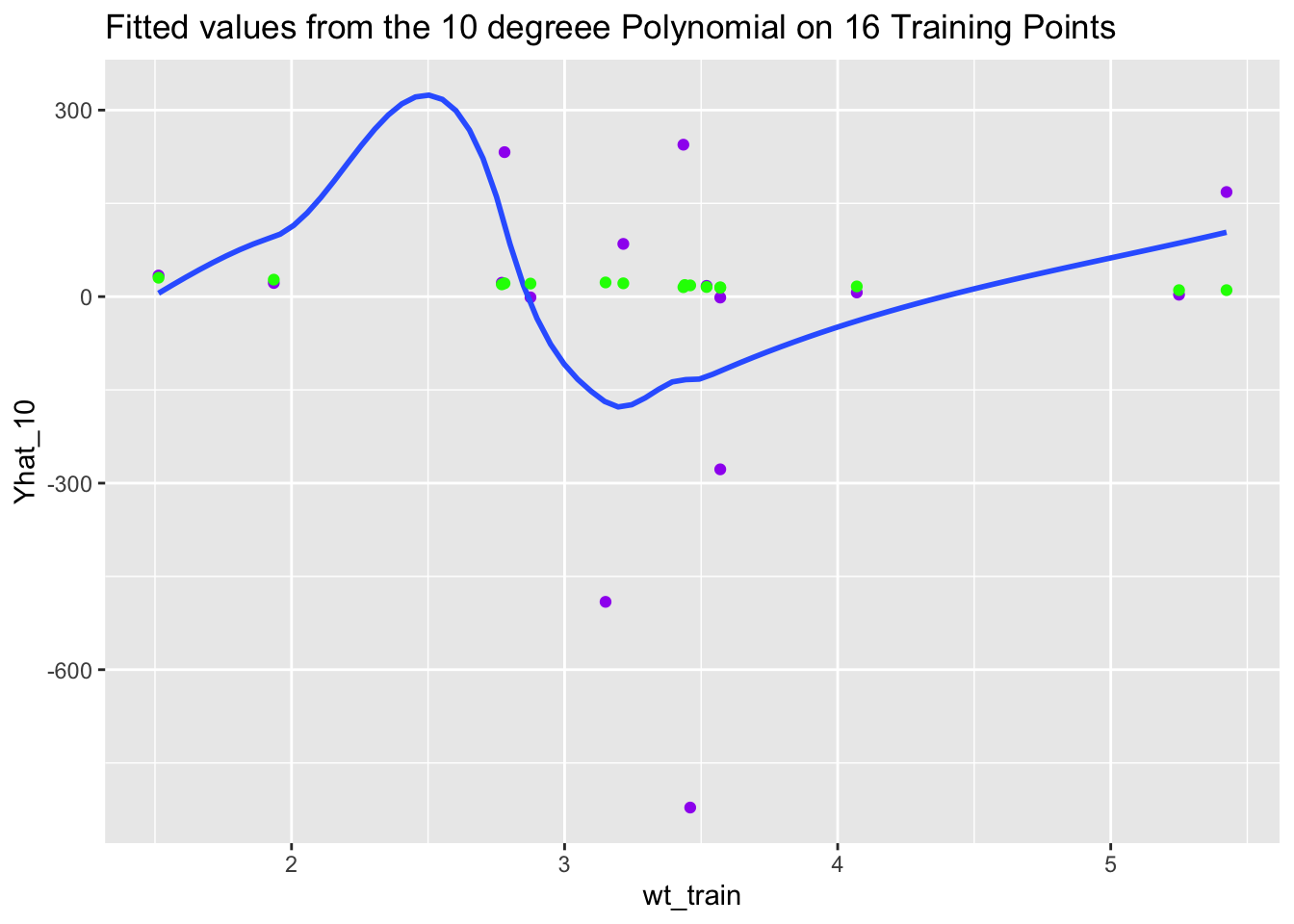
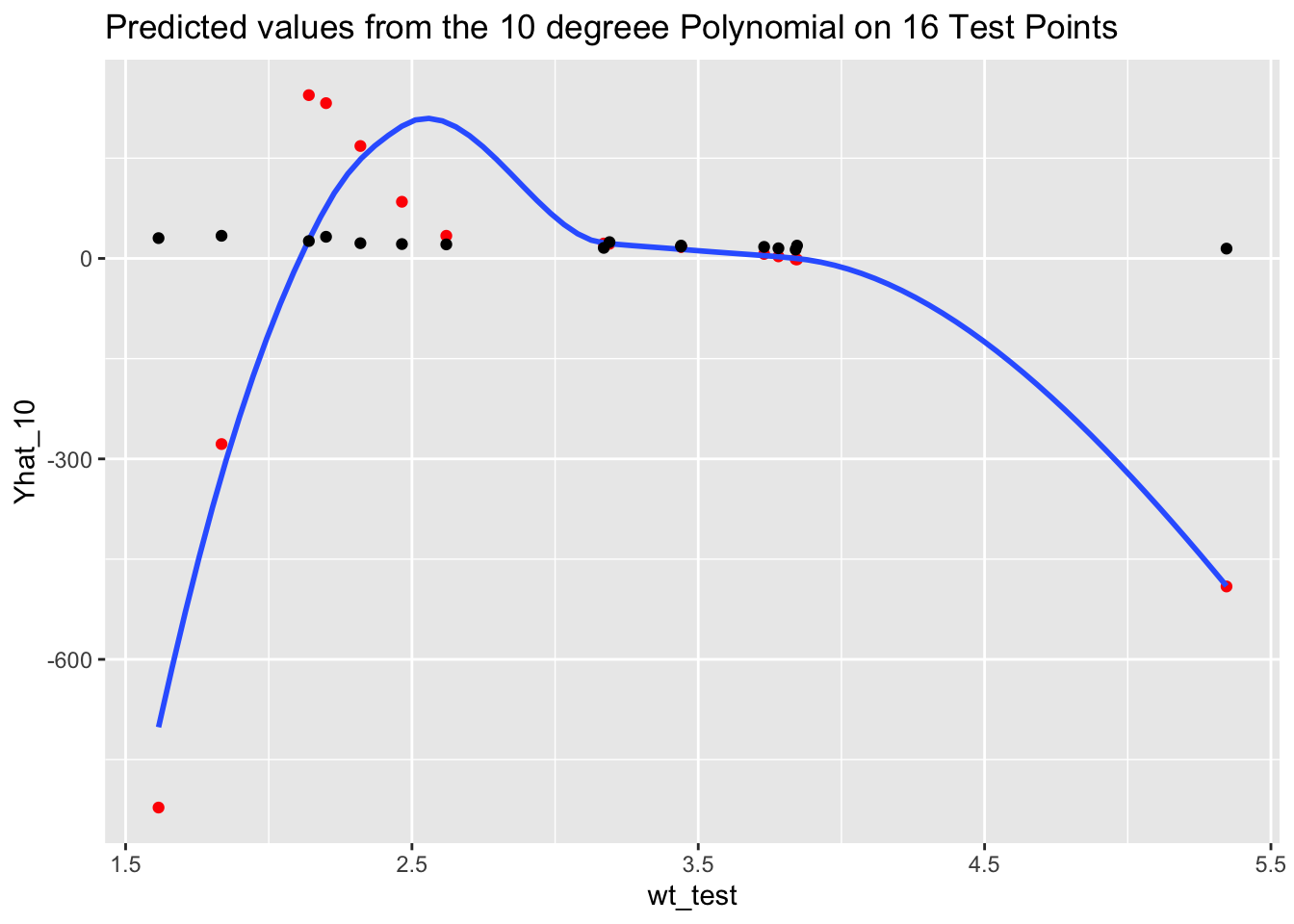
The over-fitting to noise in the training data resulted in predictions that were way off, especially in the values on either end, and thus the large RMSE for the 10th degree polynomial.
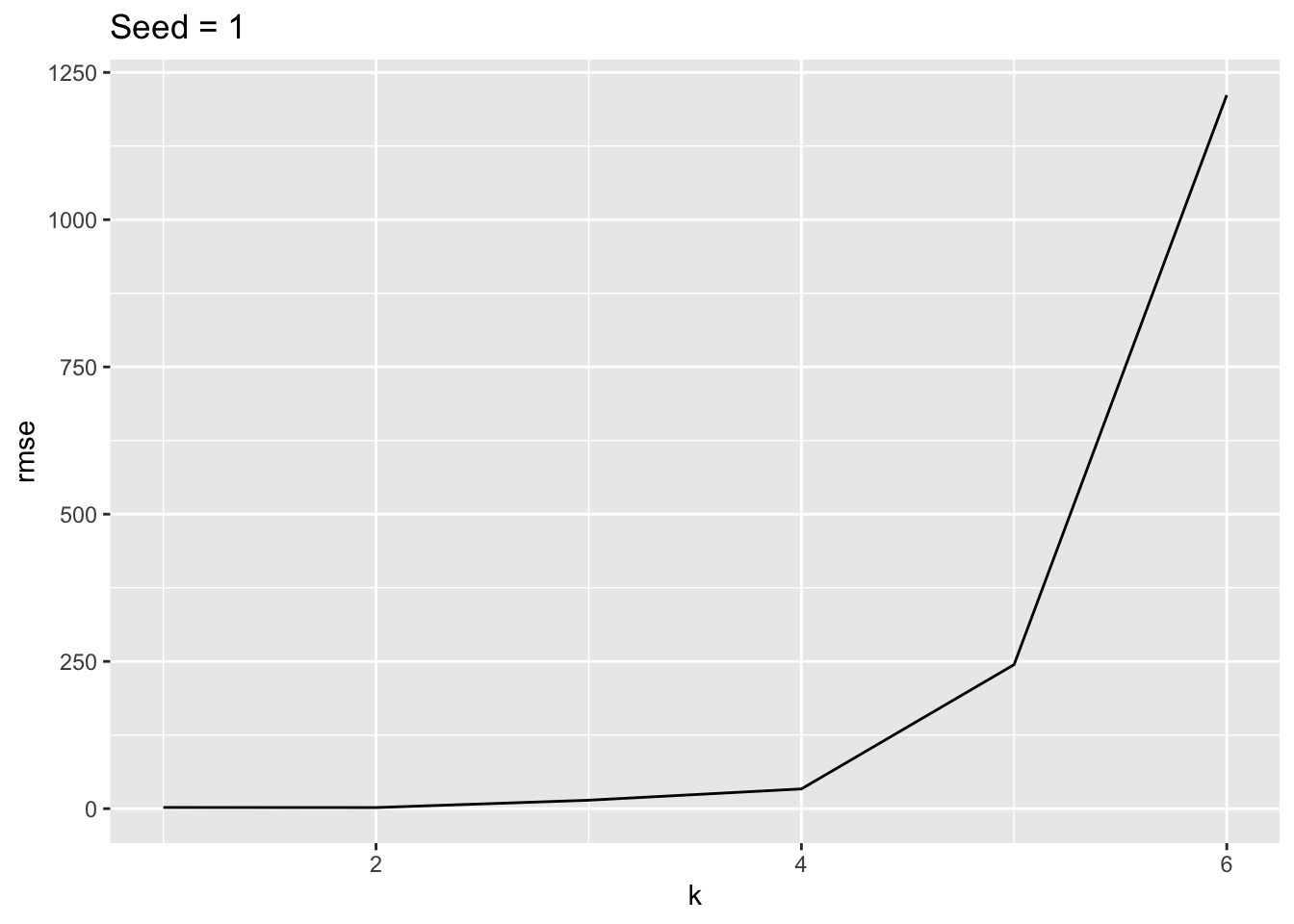
What if we plot the RMSE for the first six polynomials to better see their relative performance?
For this validation set, the 4th degree was best.
If we used a different seed to create a different split, it could be different.
set.seed(seed <- 1)
n <- nrow(mtcars)
Z <- sample(n, floor(n/2))
rmse <- vector(mode = "double", length = 10)
for (k in seq_along(rmse)) {
reg <- lm(mpg ~ poly(wt, k), data = mtcars, subset = Z)
Yhat <- predict(reg, newdata = mtcars[-Z,])
Ytest = mtcars$mpg[-Z]
rmse[[k]] <- sqrt(mean(Yhat - Ytest)^2)
}
which.min(rmse)[1] 2[1] 1.885572Now the 2nd degree polynomial is best. This is a small data set so it is not unexpected to see these changes.
Just because the predictions are off, does not mean people won’t use them.
As seen in previous example, the validation set technique uses only a portion of the data to fit the models.
\[ \widehat{\text{MSE}} = \frac{1}{n_{test}}\sum_{i \in test}(\hat{y}_i - y_i)^2 \]
This need to balance the split between training and testing data is the main disadvantage of the validation set method.
We lose a lot of information regardless of what we do.
Leave-one-out cross-validation (aka LOOCV, Jackknife) was developed by British statistician Maurice Quenouille in 1949 (he was 25 years old) and he refined it several years later.
Leave-one-out cross-validation tries to use almost as much data as possible to train the model.
Use \((n-1)\) points for training and the remaining \(n^{th}\) point for testing.
This gives almost maximum performance in fitting the model.
This is denoted as predict 1 response \(y_i\) with \(\hat{y}_{i(-i)}\) where the subscript \((-i)\) means the value \(\hat{y}_{i}\) was predicted based on a model trained without the \(i^{th}\) values of \(Y\) and \(X\) in the training data.
By itself, this is an unreliable estimate. But, we will repeat the same procedure for every data point and generate \(n\) predictions.
Then we have
\[ \widehat{\text{MSE}}_\text{jackknife} = \frac{1}{n}\sum_i^n(\hat{y}_{i(-i)} - y_i)^2 \]
Let’s repeat with mtcars for mpg.
lm().
predict() step.Yhat’s, we calculate the RMSE for the \(k\) and move to the next \(k\).We can plot the RMSE for each \(k\) as before.
We see different pattern of RMSE. Note the scale of the \(y\) axis.
We can check the \(k\) with the minimum RMSE.
We can also look at the fitted values for the 10th degree polynomial.
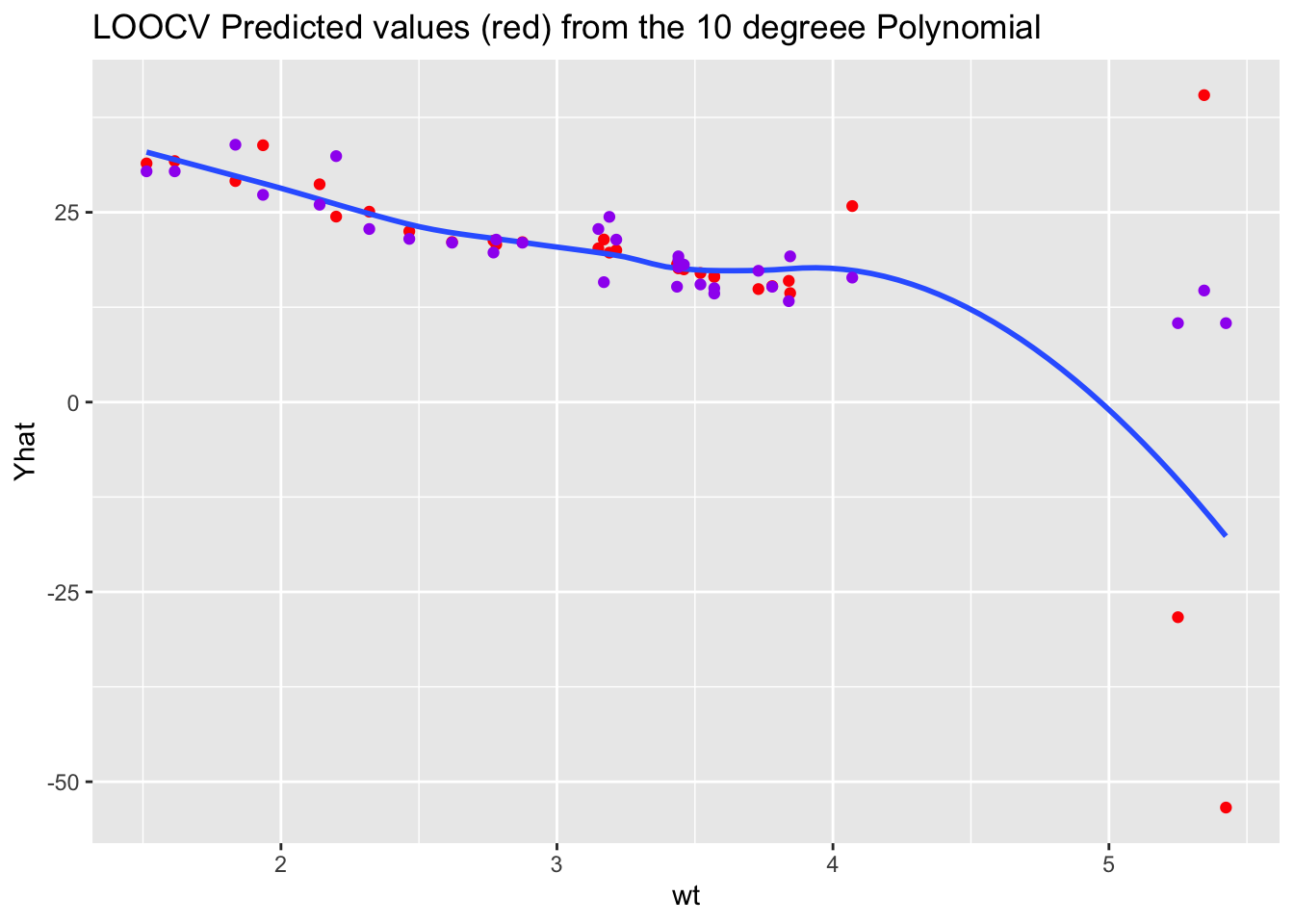
We no longer have the extreme values for the highest degree polynomial.
When we look at the chosen polynomial, we see
for (i in 1:n) { # delete one at a time
reg <- lm(mpg ~ poly(wt, 2), data = mtcars[-i ,])
Yhat[i] <- predict(reg, newdata = mtcars[i ,])
}
ggplot(bind_cols(mtcars, Yhat = Yhat), aes(wt, Yhat)) +
geom_point(color = "red") +
geom_smooth(se = FALSE) +
geom_point(mapping = aes(wt, mpg), color = "purple") +
ggtitle("LOOCV Predicted values (red) from the 2 degreee Polynomial") ->
plot_p
plot_p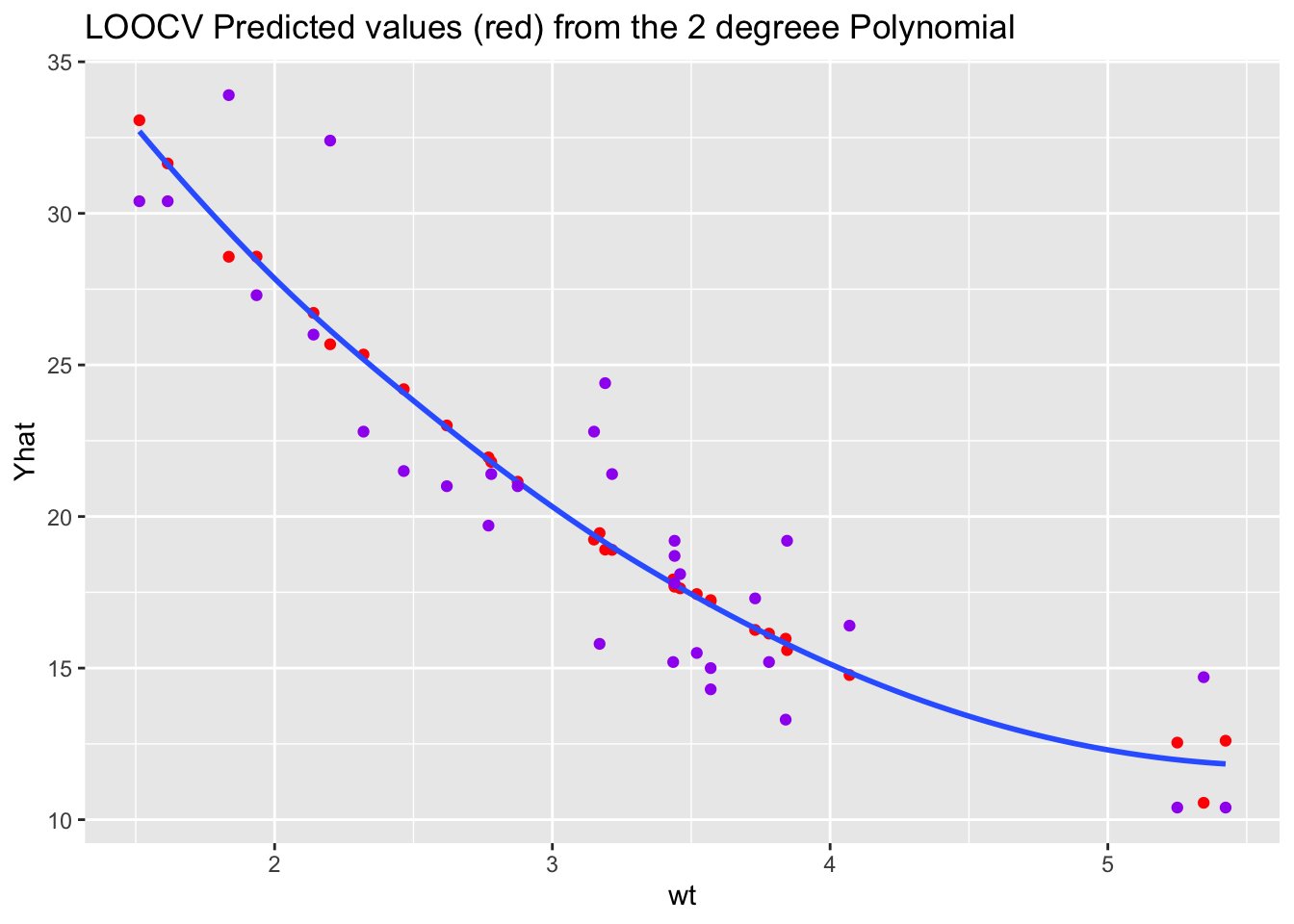
What do you notice about the \(\hat{y}\) for the recommended 2-degree polynomial?
poly() function?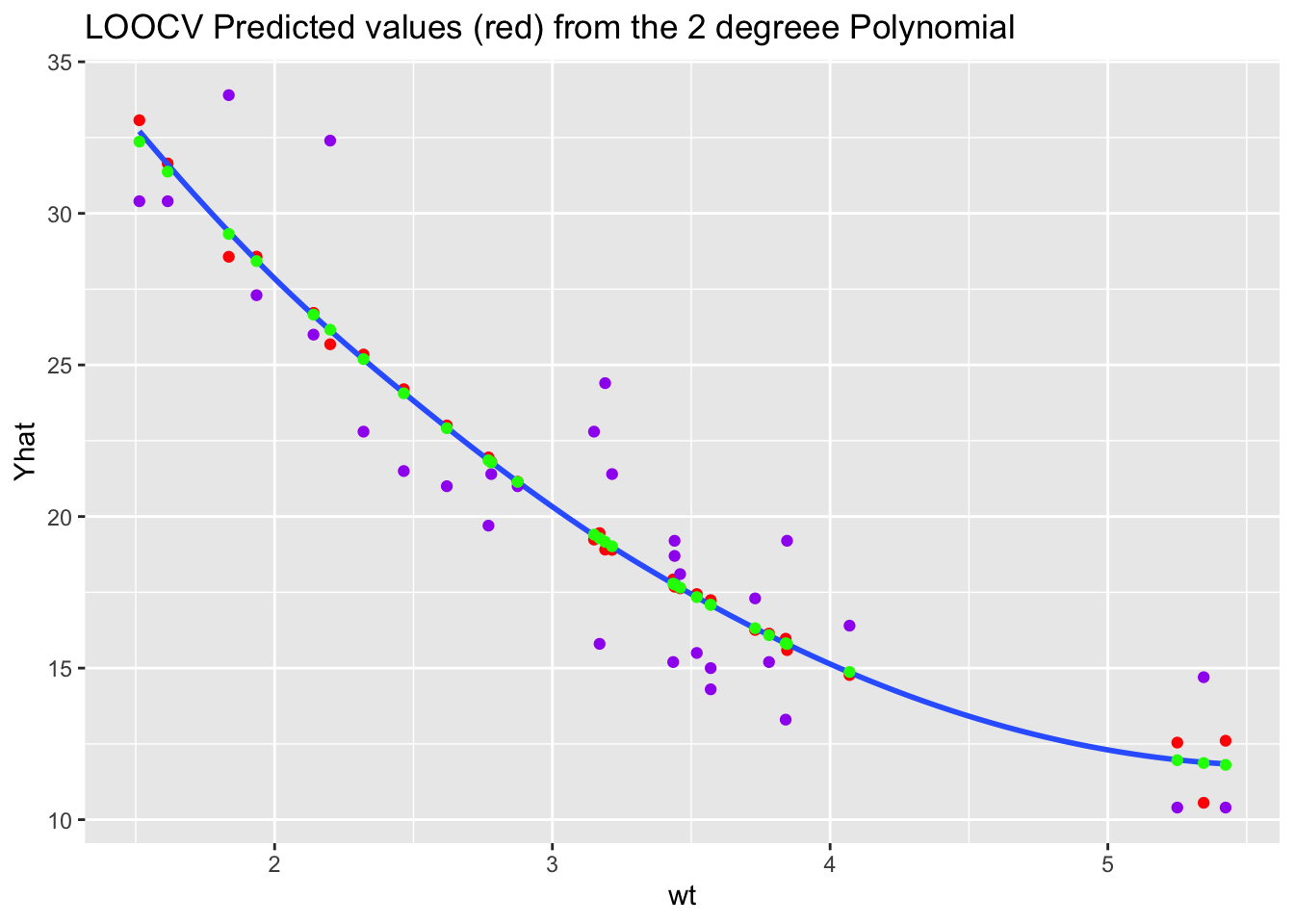
Why not?
We do not have to always write out these for loops for LOOCV. We will use the {boot} package later on which has functions we can use.
The above examples were regression problems using LOOCV for estimating the RMSE.
Earlier, the MASS::lda() function with CV = TRUE used this leave-one-out cross-validation method to produce the posterior probabilities for each category in a classification problem.
For classification, which does not have residuals that allow us to use RMSE, we can use LOOCV to estimate the prediction error rate using
\[ \frac{1}{n}\sum_{i=1}^n I_{\hat{y}_{i(-i)} \neq y_i} \]
where
\[ I_{\hat{y}_{i(-i)} \neq y_i} = \cases{ 1 \text{ if } \hat{y}_{i(-i)} \neq y_i \\ 0 \text{ otherwise }} \]
When you delete just one observation, are you really expecting the predictions to change much?
A disadvantage of LOOCV is the results lead to strongly correlated components of \(\widehat{\text{MSE}}\) since every estimate uses almost the same data.
This affects the \(Var(\widehat{\text{MSE}})\).
Consider calculating a sample mean \(\bar{x}\) to estimate the population mean \(\mu\).
\[ \text{Var}(\bar{x}) = \text{Var} \left( \frac{1}{n}\sum_{i = 1}^n x_i\right) = \frac{1}{n^2}\sum_{i = 1}^n \text{Var}(x_i) =\frac{1}{n^2}(n \sigma^2) = \frac{\sigma^2}{n} \]
So, the larger the \(n\) the smaller the variance of the estimate which makes it a more reliable estimate.
But this is only true if the \(x_1, x_2, \ldots, x_n\) are uncorrelated, i.e., \(\text{Var}(\sum x_i) = \sum \text{Var}(x_i)\) only when all covariances \(\text{Cov}(x_i, x_j) = 0 \quad \forall i\neq j\).
However, if the components of RMSE are correlated, then the covariances among the residuals are each almost as large as the variance of the residuals.
The sum is now as follows:
\[ \text{Var}(\bar{x}) = \text{Var}\sum (\hat{y}_i - y_i)^2 = \sum \text{Var}(\,\,\underbrace{(\hat{y}_i - y_i)^2}_{e_i}\,\,) + \sum_i \sum_j \text{Cov}(e_i, e_j) \ \forall (i \neq j) \tag{5.1}\]
Equation 5.1 means the \(\text{Var}(\widehat{\text{MSE}})\) for LOOCV is much larger than desired.
What is the solution?
This is our third method of cross-validation after validation set and LOOCV.
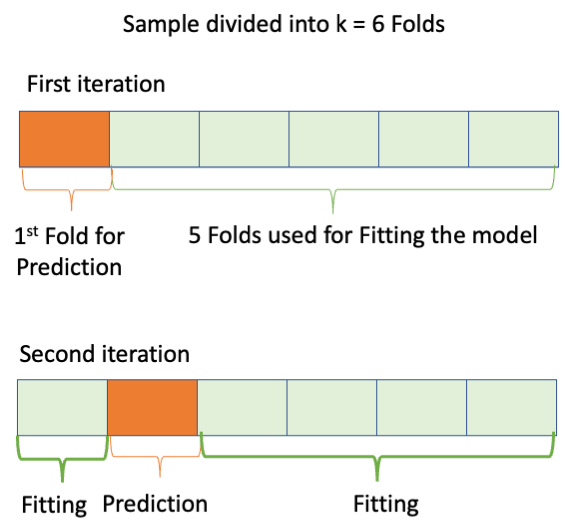
\(K\)-fold Cross-Validation deletes a group of observations (called a fold) instead of just one observation.
Given a sample of size \(n\),
We then estimate the the cross validation MSE by averaging the “K” estimates of the test error as in Equation 5.2
\[ \widehat{\text{MSE}}_{CV} = \frac{1}{n}\sum(\hat{y}_{i (-K)} - y_i)^2 \tag{5.2}\]
Equation 5.2 is equivalent to equation 5.3 in the book as seen in Equation 5.3:
\[ \text{CV}_k = \frac{1}{k}\sum_{i=1}^k\text{MSE}_i \tag{5.3}\]
You can see K-fold is a generalization of LOOCV.
Commonly used values of \(K\) are \(K=5\) and \(K=10\) based on empirical performance.
R has two packages commonly used for \(K\)-fold validation: {boot} and {bootstrap}.
Auto data set.Rows: 392
Columns: 9
$ mpg <dbl> 18, 15, 18, 16, 17, 15, 14, 14, 14, 15, 15, 14, 15, 14, 2…
$ cylinders <int> 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 4, 6, 6, 6, 4, …
$ displacement <dbl> 307, 350, 318, 304, 302, 429, 454, 440, 455, 390, 383, 34…
$ horsepower <int> 130, 165, 150, 150, 140, 198, 220, 215, 225, 190, 170, 16…
$ weight <int> 3504, 3693, 3436, 3433, 3449, 4341, 4354, 4312, 4425, 385…
$ acceleration <dbl> 12.0, 11.5, 11.0, 12.0, 10.5, 10.0, 9.0, 8.5, 10.0, 8.5, …
$ year <int> 70, 70, 70, 70, 70, 70, 70, 70, 70, 70, 70, 70, 70, 70, 7…
$ origin <int> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 3, 1, 1, 1, 3, …
$ name <fct> chevrolet chevelle malibu, buick skylark 320, plymouth sa…Calculate a linear regression of mpg on several variables using the glm function.
The glm() output object is suitable (has all the data needed) for using in a call to boot::cv.glm() which will compute the prediction error based on a \(K\)-fold cross validation of a generalized linear model.
Run the cross validation with boot::cv.glm().
Look at the help for cv.glm(). How is \(K\) chosen?
Glimpse the output.
List of 4
$ call : language cv.glm(data = Auto, glmfit = reg)
$ K : num 392
$ delta: num [1:2] 12 12
$ seed : int [1:626] 10403 20 1654269195 -1877109783 -961256264 1403523942 124639233 261424787 1836448066 1034917620 ...We see the original call and the \(K\) value (here the default).
delta is a vector of two estimates of cross-validation prediction error
How can we use this?
Let’s see if we need all the variables in the model or we can do just as well without some.
Let’s run a reduced model without displacment and cylinders.
reg2 <- glm(mpg ~ weight + year + horsepower, data = Auto)
regCV2 <- cv.glm(Auto, reg2)
regCV2$delta[1] 11.89897 11.89865This is Prediction mean squared error. What does it mean that it is smaller?
We did better without the additional variables.
Confirm with anova() with test = "Chisq".
test = "Chisq" since we are using the general linear model and our results are based on deviance which is a Chi-Squared variableAnalysis of Deviance Table
Model 1: mpg ~ weight + year + horsepower
Model 2: mpg ~ weight + year + displacement + cylinders + horsepower
Resid. Df Resid. Dev Df Deviance Pr(>Chi)
1 388 4565.7
2 386 4551.6 2 14.061 0.5509This was, by default, \(K=n\) or the same as LOOCV.
Using the same glm() object for the reduced model, we just set K = 10 which is a common practice.
delta is adjusted to reduce the bias from uneven group sizes.Just to check, set \(K=n\).
This was an example of \(K\)-fold cross validation in a regression context.
In linear regression, R was calculating the sum of the squared residuals.
For classification, we don’t have results so we use the error classification rate.
Recall
\[ \widehat{\text{Error rate}} = \frac{1}{n}\sum I_{\hat{y}_{i(-i)} \neq y_i} \]
To do this, we will use a suitable Loss function.
Let’s define a Loss function \(L\) of the actual response value \(y\) and the estimated probability \(p\).
\[ \text{L}(y, p) = \cases{1 \qquad \text{if }\, y = 1 \text{ but }p \leq \text{threshold} \implies \hat{y} = 0 \\ \qquad \,\,\text{or }y = 0 \text{ but } p > \text{threshold}\implies \hat{y} = 1\\ \\ 0 \qquad \text{otherwise (we had correct prediction)}} \tag{5.4}\]
We can define Equation 5.4 as a function in R and use in glm() for logistic regression where we are predicting probabilities.
As an example, assume actual \(y_i= 1\) and an estimated \(p = .9\), the loss is
However, if actual \(y_i= 1\) and an estimated \(p = .3\), the loss is
Since we included mean() in our loss function, we can use a vector of predictions:
Let’s load the Titanic data set from before.
titanic <- readr::read_csv("https://raw.githubusercontent.com/rressler/data_raw_courses/main/titanic_dalex.csv")
glimpse(titanic)Rows: 2,207
Columns: 9
$ gender <chr> "male", "male", "male", "female", "female", "male", "male", "…
$ age <dbl> 42.0000000, 13.0000000, 16.0000000, 39.0000000, 16.0000000, 2…
$ class <chr> "3rd", "3rd", "3rd", "3rd", "3rd", "3rd", "2nd", "2nd", "3rd"…
$ embarked <chr> "Southampton", "Southampton", "Southampton", "Southampton", "…
$ country <chr> "United States", "United States", "United States", "England",…
$ fare <dbl> 7.1100, 20.0500, 20.0500, 20.0500, 7.1300, 7.1300, 24.0000, 2…
$ sibsp <dbl> 0, 0, 1, 1, 0, 0, 1, 1, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0…
$ parch <dbl> 0, 2, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 0, 0, 0, 0, 0, 0…
$ survived <chr> "no", "no", "no", "yes", "yes", "yes", "no", "yes", "yes", "y…Let’s check if embarked is a useful predictor variable.
survived to 0 or 1.
no yes
1496 711
0 1
1496 711 NAsNow do the LOOCV and get the deltas.
Now use the loss function.
Since we used our Loss function, this is now the classification error rate.
We can set \(K\) as well.
Let’s fit a model without embarked and compare.
log_reg <- glm(Y ~ gender + age + class + fare,
data = titanic2, family = "binomial")
set.seed(123)
cv.glm(titanic2, log_reg, cost = L, K = 10)$delta[1] 0.2371429 0.2367143Slightly lower error rate.
What if I delete class?
log_reg <- glm(Y ~ gender + age + fare,
data = titanic2, family = "binomial")
set.seed(123)
cv.glm(titanic2, log_reg, cost = L, K = 10)$delta[1] 0.2585714 0.2598571Adjusted error rate went up so there is some value in class.
You can use cross validation to compare models for any kind of tuning.
Jackknife is a method for reducing bias. It was developed in 1949 as mentioned earlier.
Since MSE is the sum of Bias and Variance, we would like to reduce the Bias without increasing the Variance.
If \(E[\hat{\theta}] = \theta\) then we say \(\hat{\theta}\) is unbiased estimator of \(\theta\).
Under the standard linear regression assumptions, our estimators for the \(\beta\)s are unbiased.
However, suppose \(\theta\) is the population maximum. If we have a \(\hat{\theta}\) from a sample, is it unbiased?
Assume we have some population statistic, \(\theta\), which is unknown, and we want to estimate it with an estimator \(\hat{\theta}\).
Let’s define the bias of \(\hat{\theta}\) as:
\[ \text{Bias}(\hat{\theta}) = E[\hat{\theta}] - \theta \tag{5.5}\]
We can collect multiple samples from a population and each sample will get its own \(\hat{\theta}_i\).
The expected value of \(\hat{\theta}\) (denoted by \(E[\hat{\theta}]\)) is the average of all possible \(\hat{\theta}_i\) from all possible samples of our population.
We would love for \(E[\hat{\theta}]\) to be \(\theta\).
However, for many \(\theta\), there is no unbiased estimator for a given population.
The sample proportion \(\hat{p}\) is a unbiased estimator for the parameter \(p\) of a Binomial distribution
However, the logit of the proportion, \(\text{logit}(\hat{p}) = \ln(\hat{p}/(1-\hat{p}))\), is a nonlinear function of the proportion \(\hat{p}\) and we can show it must be biased.
\[ p(X_1, X_2, \ldots X_n) = \prod p(X_i) = p^{\sum X_i}(1-p)^{(n-\sum X_i)} \rightarrow \text{a polynomial of }p \]
\[ E[\hat{\theta}] = \sum\underbrace{\hat{\theta}(X_1, X_2, \ldots X_n)}_{\text{Does not depend on }p}\,* \underbrace{p(X_1, X_2, \ldots X_n)}_{\text{a polynomial of }p} \tag{5.6}\]
Therefore, Equation 5.6 shows the expected value of an estimator, \(\hat{\theta}\), of any statistic of a population with a binomial distribution, can only be a polynomial.
Since the logit function \(\text{logit}(p) = \ln(p/(1-p))\) is Not a polynomial, it follows that
Since our goal is reducing Bias + Variance, sometimes, even when there is an unbiased estimator, we might prefer using a biased estimator with other nice properties such as reducing the MSE.
When we are using a biased estimator, we can use Jackknife as a method to reduce the bias.
In 1989, Doss and Sethuraman proved the following caution about bias reduction:
If there is no unbiased estimator \(\hat{\theta}\), but you can construct a sequence of estimators such that \(\text{Bias}(\hat{\theta}) \rightarrow 0\), i.e., using repeated jackknifes, \(\text{Variance}(\hat{\theta}) \rightarrow \infty\).
Quenouille noticed the bias of an estimator tends to reduce as the sample size \(n\) increases.
That suggested using a power series to represent the unknown function for the bias of \(\hat{\theta}\):
\[ \text{Bias}(\hat{\theta}) = \overbrace{\frac{a_1}{n} + \frac{a_2}{n^2} + \frac{a_3}{n^3} + \cdots}^{\text{the Bias}} \qquad \implies \text{of order} \frac{1}{n} \tag{5.7}\]
where \(a_1, a_2, a_3, \ldots\) are some coefficients that are unknown.
We use Big \(O\) notation to represent what happens to the results as \(n \rightarrow \infty\).
Here it means that as \(n \rightarrow \infty\), the \(a_1/n\) term will be the largest term, regardless of the value of any \(a_i\), as the other terms will approach 0 much faster.
We can then use Equation 5.5 to express the expected value of \(\hat{\theta}\) as \(\theta\) + the bias, or,
\[ E[\hat{\theta}]=\theta + \overbrace{\frac{a_1}{n} + \frac{a_2}{n^2} + \frac{a_3}{n^3} + \cdots }^{\text{the Bias}} \tag{5.8}\]
Now let’s apply jackknife, a LOOCV approach.
\[ \text{Average: }\equiv \,\hat{\theta}_{(.)}= \frac{1}{n} \sum_{i=1}^n \hat{\theta}_{(-i)} \]
That gives us an expected value of:
\[ E[\hat{\theta}_{(.)}]=\theta + \overbrace{\frac{a_1}{n-1} + \frac{a_2}{(n-1)^2} + \frac{a_3}{(n-1)^3} + \cdots }^{\text{the Bias}} \tag{5.9}\]
Let’s look at Equation 5.8 and Equation 5.9 together and see if we can reduce the variance by combining them in a way that eliminates the first term of the bias, the \(O(n)\) term.
Start with the following
\[ \begin{align} E[\hat{\theta}] & = \theta + \quad\frac{a_1}{n} \,\,\,+ \quad\,\frac{a_2}{n^2} \quad\,\,\, +\quad \, \frac{a_3}{n^3} \quad \,+ \cdots \\ \\ E[\hat{\theta}_{(.)})]&= \theta + \,\frac{a_1}{n-1} + \frac{a_2}{(n-1)^2} + \frac{a_3}{(n-1)^3} + \cdots \end{align} \]
Multiply the first equation by \(n\) on both sides and the second equation by \((n-1)\) on both sides.
\[ \begin{align} n*E[\hat{\theta}] & = \,\,\,\,\,\,n\theta \quad \,\,+ \frac{a_1}{1} \,\,\,+ \quad\,\frac{a_2}{n} \quad\, +\quad \, \frac{a_3}{n^2} \quad \,+ \cdots \\ \\ (n-1)*E[\hat{\theta}_{(.)})]&= (n-1)\theta + \,\frac{a_1}{1} + \frac{a_2}{(n-1)^1} + \frac{a_3}{(n-1)^2} + \cdots \end{align} \tag{5.10}\]
Now subtract the bottom of Equation 5.10 from the top:
\[ nE[\hat{\theta}] - (n-1)E[\hat{\theta}_{(.)}] = E[n\,\hat{\theta} - (n-1)\hat{\theta}_{(.)}] \]
\[ \begin{align} E[n\,\hat{\theta} - (n-1)\hat{\theta}_{(.)}] & = \bigg(n\theta - (n-1)\theta\bigg) + (a_1 - a_1) + \left(\frac{a_2}{n} - \frac{a_2}{(n-1)}\right) + \cdots\\ \\ & = \theta + 0 +\frac{(n-1)a_2 - na_2}{n(n-1)} + \cdots\\ \\ &= \theta + \quad \frac{-a_2}{n^2-n} \,\,\,+ \cdots \\ \\ E[\hat{\theta}_{JK}] =E[ n\hat{\theta} - (n-1)\hat{\theta}_{(.)}]&= \theta + \qquad\, \frac{b_2}{n^2}\quad\,+\cdots \qquad \implies \text{of order } \frac{1}{n^2} << \frac{1}{n} \end{align} \tag{5.11}\]
So the bias has been reduced as the \((1/n^2)\) will be smaller in magnitude than the \(1/n\) terms from Equation 5.7.
The left side of Equation 5.11 is called the Jackknife Estimator, \(\hat{\theta}_{JK}\) and we will use the following formula for it to reduce the bias of an original estimate.
\[ \hat{\theta}_{JK}= n\hat{\theta} - (n-1)\hat{\theta}_{(.)} \tag{5.12}\]
As seen in Equation 5.11, \(\hat{\theta}_{JK}\) has a smaller bias than the original \(\hat{\theta}\).
Bias may be positive or negative.
As a result, \(\hat{\theta}_{JK}\) may be smaller or larger than the original \(\hat{\theta}\) to correct the over-estimation or under-estimation of the original estimator.
Example: If we have a sample size of \(1000\), then \(\hat{\theta}\) has a bias of order \(b/1000\) and \(\hat{\theta}_{JK} = n\hat{\theta} - (n-1)\hat{\theta}_{(.)}\) has a bias of order \(b/(1000)^2\).
The method has three steps once you have calculated the original (potentially-biased) estimator \(\hat{\theta}\) using all \(n\) values in the sample.
We want to estimate the height of the tallest person on earth (in inches)
Let’s apply Jackknife
Now calculate the average: \(\hat{\theta}_{.} = \frac{72 + 72 + 70 + 72}{4} = 71.5\)
Calculate the jackknife estimate. \(n = 4\) so
\[ \begin{align} \hat{\theta}_{JK} &= n\,\hat{\theta} - (n-1) \hat{\theta}_{(.)} \\ &= 4 * 72 - 3 * 71.5 \\ &= 288 - 214.5 \\ &= 73.5 \end{align} \]
Note. It is still biased but it has less bias than before.
Let’s estimate the amount of bias of the original \(\hat{\theta}\).
\[ \begin{align} \widehat{\text{Bias}}(\hat{\theta})&= \hat{\theta} - \hat{\theta}_{JK} \\ &= 72 - 73.5 \\ &= -1.5 \end{align} \]
bootstrap::jackknife() to Estimate the Maximum of mtcars$mpg.We will use the jackknife() function from the {bootstrap} package for Jackknife methods.
bootstrap::jackknife() requires us to define a function to calculate our estimator \(\theta\) since we can use the jackknife for any kind of estimator we want.
Let’s create a function to find the maximum of a sample.
Let’s look at mpg in the mtcars data set.
Min. 1st Qu. Median Mean 3rd Qu. Max.
10.40 15.43 19.20 20.09 22.80 33.90 [1] 32Use the bootstrap::jackknife() function with the data and the my_max function for \(\theta\).
my_max and not calling it using the my_max().$jack.se
[1] 1.453125
$jack.bias
[1] -1.453125
$jack.values
[1] 33.9 33.9 33.9 33.9 33.9 33.9 33.9 33.9 33.9 33.9 33.9 33.9 33.9 33.9 33.9
[16] 33.9 33.9 33.9 33.9 32.4 33.9 33.9 33.9 33.9 33.9 33.9 33.9 33.9 33.9 33.9
[31] 33.9 33.9
$call
jackknife(x = mtcars$mpg, theta = my_max)The jack.values are the 32 estimates \(\hat{\theta}_{(-i)}\) with one value deleted.
Compare them to the original values.
32.4 33.9
1 31
10.4 13.3 14.3 14.7 15 15.2 15.5 15.8 16.4 17.3 17.8 18.1 18.7 19.2 19.7 21
2 1 1 1 1 2 1 1 1 1 1 1 1 2 1 2
21.4 21.5 22.8 24.4 26 27.3 30.4 32.4 33.9
2 1 2 1 1 1 2 1 1 Let’s get the mean \(\hat{\theta}_{(.)}\) and then calculate the jackknife estimate using Equation 5.12.
n <- nrow(mtcars)
jk_dot <- mean(jk$jack.values)
theta_jk <- n * max(mtcars$mpg) - (n - 1) * jk_dot
theta_jk [1] 35.35313Now we can get the bias.
Or we can calculate the original \(\hat{\theta}\) and subtract the bias to get the jackknife estimate.
mtcars$mpg.We can calculate the variance of a sample using the R var() function.
Let’s define a function to calculate a sample variance based on the definition of a population variance:
Why do we get a different result?
The standard estimator of variance is
\[ \hat{\sigma}^2 = s^2 = \frac{1}{n-1}\sum_i^n(x_i - \bar{x})^2 \]
This is what R uses in var() as it divides by \((n-1)\) to make the output unbiased.
In my formula we used the mean() which means we used \(1/n\) so my_var() was biased as it underestimated the variance.
Let’s calculate the jackknife output for my_var().
$jack.se
[1] 8.75157
$jack.bias
[1] -1.135128
$jack.values
[1] 36.29657 36.29657 36.07967 36.26701 36.25971 36.19215 35.20755 35.70572
[9] 36.07967 36.29769 36.14939 35.87055 36.06479 35.52766 33.19709 33.19709
[17] 35.35648 31.27867 32.78502 29.97409 36.25796 35.62237 35.52766 34.78862
[25] 36.29769 34.59340 35.16129 32.78502 35.71109 36.31902 35.46119 36.26701
$call
jackknife(x = mtcars$mpg, theta = my_var)Let’s take the original estimate and subtract the bias calculated by jackknife():
We get the same value as the unbiased estimator.
The jackknife method allows us to create an unbiased estimate.
We want to use data from a vaccine trial to estimate its effectiveness in terms of the probability of creating antibodies after the vaccine.
Say we have a sample of 20 and the first \(X_1, X_2, \ldots, X_{13} = 1\) and \(X_{14}, X_{15}, \ldots, X_{20} = 0\)
We use the sample proportion to estimate \(p\) so we estimate \(\hat{p} = 13/20\). This is a unbiased as it is also a sample mean (linear combination of the samples).
We want to estimate the logit of \(p\), \(\ln(\frac{p}{1-p})\), which is a non-linear function of \(p\).
That means \(\widehat{\text{Logit}}(p) = \ln\left(\frac{\hat{p}}{(1-\hat{p})}\right)\) may be biased.
You may have heard of Jensen’s inequality which states several properties about the expectation of a function \(g(x)\).
\[ \begin{align} 1. \qquad E[g(x)] &\geq g(E[x]) \iff \quad g(x) \text{ is convex} \iff \text{second derivative is postive: smile} \implies \text{Overestimates}\\ \\ 2. \qquad E[g(x)] &\leq g(E[x]) \iff \quad \text{ is concave} \iff \text{second derivative is negative: frown}\implies \text{Underestimates}\\ \\ 3. \qquad E[g(x)] &= g(E[x]) \iff \quad \text{ is linear} \iff \text{second derivative} = 0\implies \text{Unbiased} \end{align} \tag{5.13}\]
Equation 5.13 shows why the sample mean is unbiased, as it is a linear function of the sample values.
For our sample standard deviation, the \(\sqrt{}\) function is concave. Thus case 2 from Equation 5.13 applies and we have:
\[ E[s] = E[\sqrt{s^2}]\quad \leq \quad \sqrt{E[s^2]} = \sqrt{\sigma^2} = \sigma \]
This means the standard deviation underestimates the population standard deviation \(\sigma\).
We saw that we could use jackknife to correct for this bias.
What about the logit function? Is it Convex or Concave?
\[ \begin{align} \text{Logit}(p) &= \ln\bigg(\frac{p}{(1-p)}\bigg) = \ln(p) - \ln(1-p)) \\ \\ \text{first derivative } \text{Logit}'(p)&=\,\,\,\,\, \frac{1}{p} - \frac{-1}{1-p} = \frac{1}{p} + \frac{1}{1-p}\\ \\ \text{second derivative }\text{Logit}''(p) &= \frac{-1}{p^2} + \frac{1}{(1-p)^2} \end{align} \]
This will be positive or negative depending upon the value of \(p > .5\).
suppressMessages(library(tidyverse))
df <- tibble(p = rep(1:1000)/1000,
y = log(p/(1 - p))
)
ggplot(df, aes(p, y)) +
geom_line() +
annotate("text", x = .7, y = 3, label = "Convex\n so Overestimates", size = 5) +
annotate("text", x = .25, y = -3, label = "Concave\n so Underestimates", size = 5) +
geom_vline(xintercept = .5, color = "red", lty = 2) +
ylab(latex2exp::TeX("Logit$(p) = \\frac{p}{(1-p)}$"))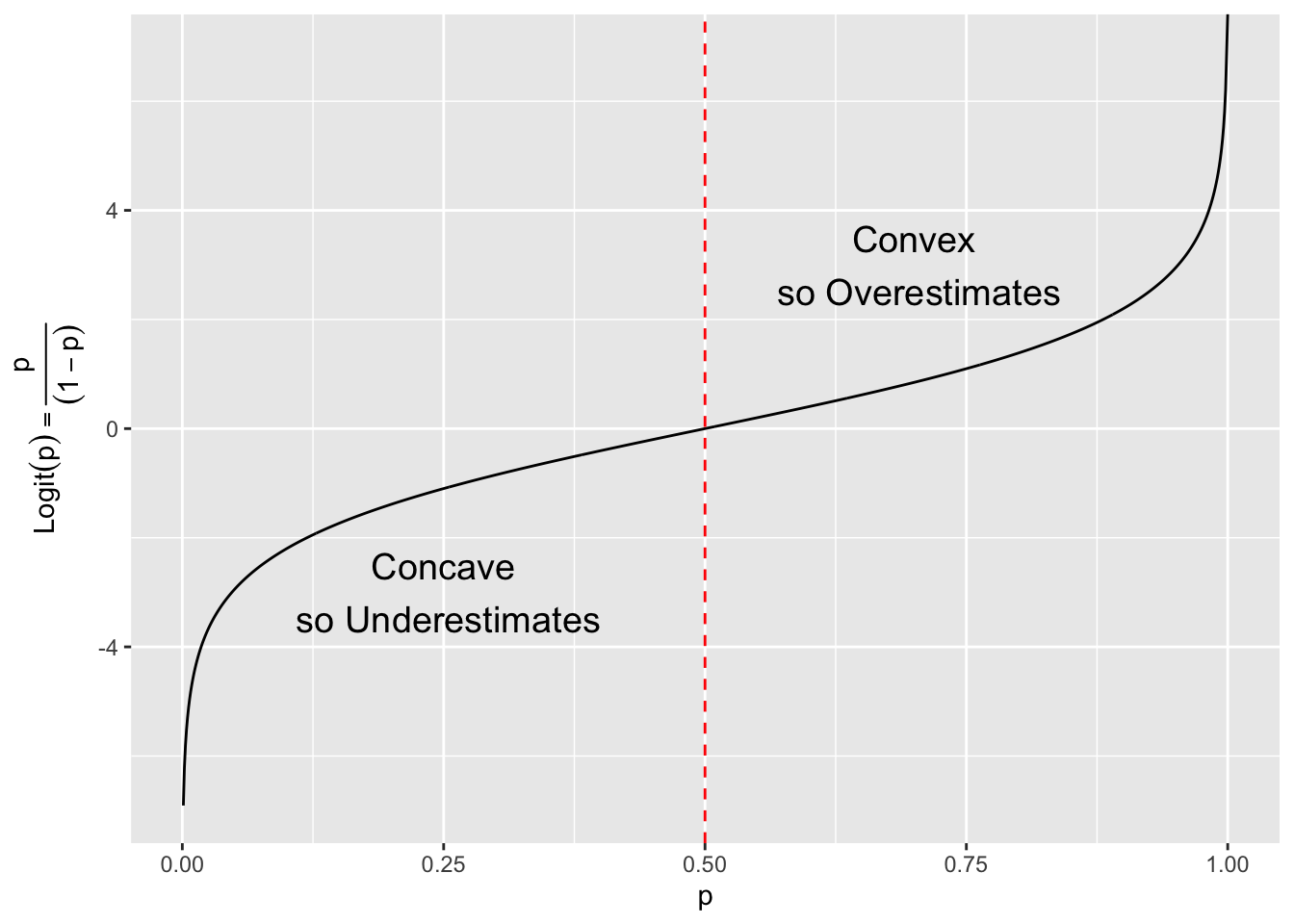
Let’s go through the Jackknife steps:
Step 1: Delete 1 at a time and calculate the estimate \(\hat{\theta}_{(-1)}, \ldots, \hat{\theta}_{(-n)}\)
Step 2: Average the results: \(\hat{\theta}_{(.)} = \frac{1}{n} \sum \hat{\theta}_{(-i)}\)
Step 3: Apply the jackknife formula \(\hat{\theta}_{JK} = n\hat{\theta} - (n-1) \hat{\theta}_{(.)}\)
Step 1: We delete one at a time and we get 20 estimates of the sample logit. With our sample of 13 1’s and 7 0’s we can simplify:
Step 2: Average
\[ \hat{\theta}_{(.)} = \frac{13 * \ln(12/7) + 7 * \ln(13/6)}{20} = 0.621 \]
Step 3. The original \(\hat{p}\) was 0.65 with \(\text{Logit}(\hat{p}) =\) 0.6190392, so we use Equation 5.12 to get
\[ \hat{\theta}_{JK} =20 * \ln(.65/.35) - 19*.621 = 0.582 \]
To estimate the bias of \(\hat{\theta}\), use \(\hat{\theta} - \hat{\theta}_{JK} = 0.619 - .582= .037\)
Conclusion: for this \(p\) the sample logit overestimates the true logit and our new estimate should be less biased.
Bootstrap is another method of evaluating the performance of a model/method and generating confidence intervals.
Let’s say we want to know the \(\text{Var}(\hat{\theta})\) for some \(\theta\) for the population.
Bootstrap replaces the population with our sample and then generates (all possible) a large number of samples of size \(n\) by sampling with replacement.
We compute our statistics from each sample.
We then use the \(n\) estimates to get a bootstrap distribution of \(\hat{\theta}\).
One can use the distribution of \(\hat{\theta}\) to create a bootstrap confidence interval for the true value of \(\theta\).
We want to calculate the properties of the sample median for mtcars$mpg.
We’ll use 10,000 samples.
Let’s generate the samples and calculate the median for each sample
[1] 19.20 18.95 19.20 19.70 21.00 18.65 20.35 17.80 20.10 18.95We can now plot the distribution of our bootstrap estimates of the median.
We can calculate various statistics as well.
[1] 19.28492[1] 19.2[1] 1.262394[1] 1.59363897.5%
21.4 To calculate confidence intervals:
2.5% 97.5%
16.8 21.4 0.5% 99.5%
15.8 22.1 ggplot(tibble(bootstrap_medians), aes(bootstrap_medians)) +
geom_histogram(bins = 50) +
geom_vline(xintercept = quantile(bootstrap_medians, probs = c(.025)),
color = "red", lty = 2) +
geom_vline(xintercept = quantile(bootstrap_medians, probs = c(.975)),
color = "red", lty = 2) +
geom_vline(xintercept = quantile(bootstrap_medians, probs = c(.005)),
color = "green", lty = 2) +
geom_vline(xintercept = quantile(bootstrap_medians, probs = c(.995)),
color = "green", lty = 2)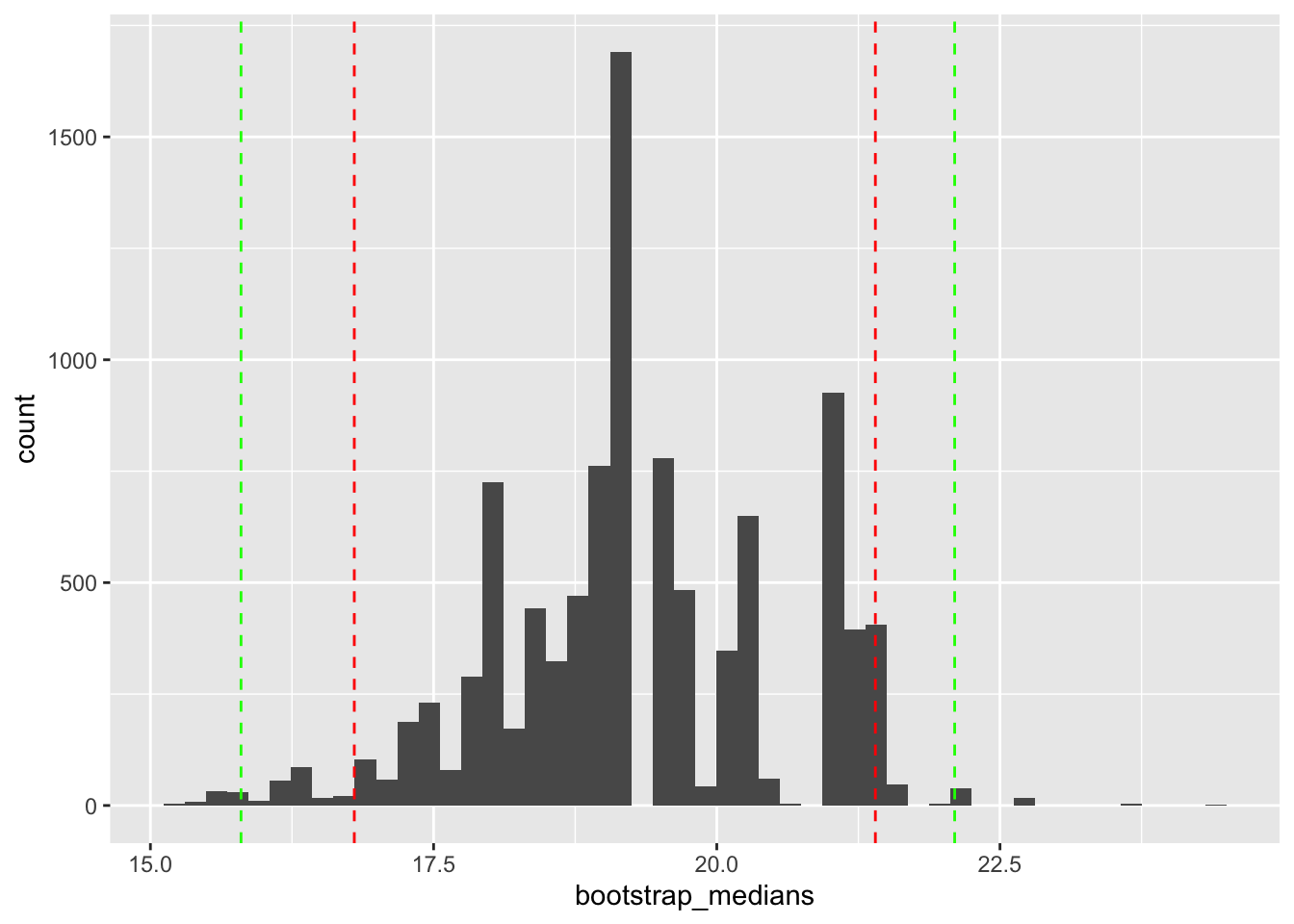
This has the same interpretation as usual.
This is the same package we used for cross-validation.
We again have to define a function for our estimator and that function must have two arguments:
Let’s define a function for the median of a sample.
We can compare our function to the R median function using the entire sample.
This only gives us one sample estimate of the sample median. We can’t calculate any statistics about our estimator with one sample.
Now set up to run with bootstrap samples
[1] 19.2Let’s use boot::boot() to generate our 10,000 bootstrap samples
ORDINARY NONPARAMETRIC BOOTSTRAP
Call:
boot::boot(data = mtcars$mpg, statistic = my_median, R = 10000)
Bootstrap Statistics :
original bias std. error
t1* 19.2 0.0881 1.272659# A tibble: 1 × 3
statistic bias std.error
<dbl> <dbl> <dbl>
1 19.2 0.0881 1.27The more samples, the more the estimate converges to the maximum likelihood estimate.
This is a non-parametric method. We have not made any assumptions about the distribution of \(\hat{\theta}\).
The content of b_out has several elements in the list
[1] "boot"[1] 19.2 [1] 21.40 19.85 20.35 20.10 20.35 16.10 21.00 18.40 17.95 18.40function(x, subsample) median(x[subsample], na.rm = TRUE)
<bytecode: 0x10af431f8>What if we want the inter-quartile range?
The clinical trial include 44,000 participants, randomized into two equal groups.
Result - 170 participants contacted Covid-19.
One can claim from these results that the vaccine reduces the chance of infection by a factor of \(20=\frac{162}{8}\).
Let’s use this ratio as our estimate of interest, \(\hat{\theta}\).
\[ \hat{\theta} = \frac{\text{count placebo patients infected}}{\text{count vaccine patients infected}} \]
Critics claimed that with only 170 cases the results were not valid.
Let’s calculate a 95% confidence interval to give a measure of the uncertainty associated with the “single number” \(\hat{\theta}\).
Create a data frame of the trial data of patients and cases.
patients = 1 means vaccine, and,cases = 1 means infectedtrial <- data.frame(
patients = c(rep(1, 22000), rep(0, 22000)), # patients (vaccine, placebo)
cases = c(rep(1, 8), rep(0, 21992), rep(1, 162), rep(0, 21838))
)
head(trial) patients cases
1 1 1
2 1 1
3 1 1
4 1 1
5 1 1
6 1 1Define the function for our estimate \(\hat{\theta}.\)
Let’s get the overall sample estimate and try a sub sample.
We can see a range of results.
Let’s compare across different number of samples.
ORDINARY NONPARAMETRIC BOOTSTRAP
Call:
boot::boot(data = trial, statistic = ratio, R = 100)
Bootstrap Statistics :
original bias std. error
t1* 20.25 3.865687 11.1073
ORDINARY NONPARAMETRIC BOOTSTRAP
Call:
boot::boot(data = trial, statistic = ratio, R = 1000)
Bootstrap Statistics :
original bias std. error
t1* 20.25 Inf NaN
ORDINARY NONPARAMETRIC BOOTSTRAP
Call:
boot::boot(data = trial, statistic = ratio, R = 10000)
Bootstrap Statistics :
original bias std. error
t1* 20.25 Inf NaNNAN’s or inf in the results for bias and standard error means some samples had 0 in the denominator since the sample didn’t select any of the positive cases.Let’s plot the histogram of our 10,000 samples:
suppressMessages(library(ggplot2))
ggplot(data.frame(thetaHat = b_out$t), aes(thetaHat)) +
geom_histogram(bins = 50)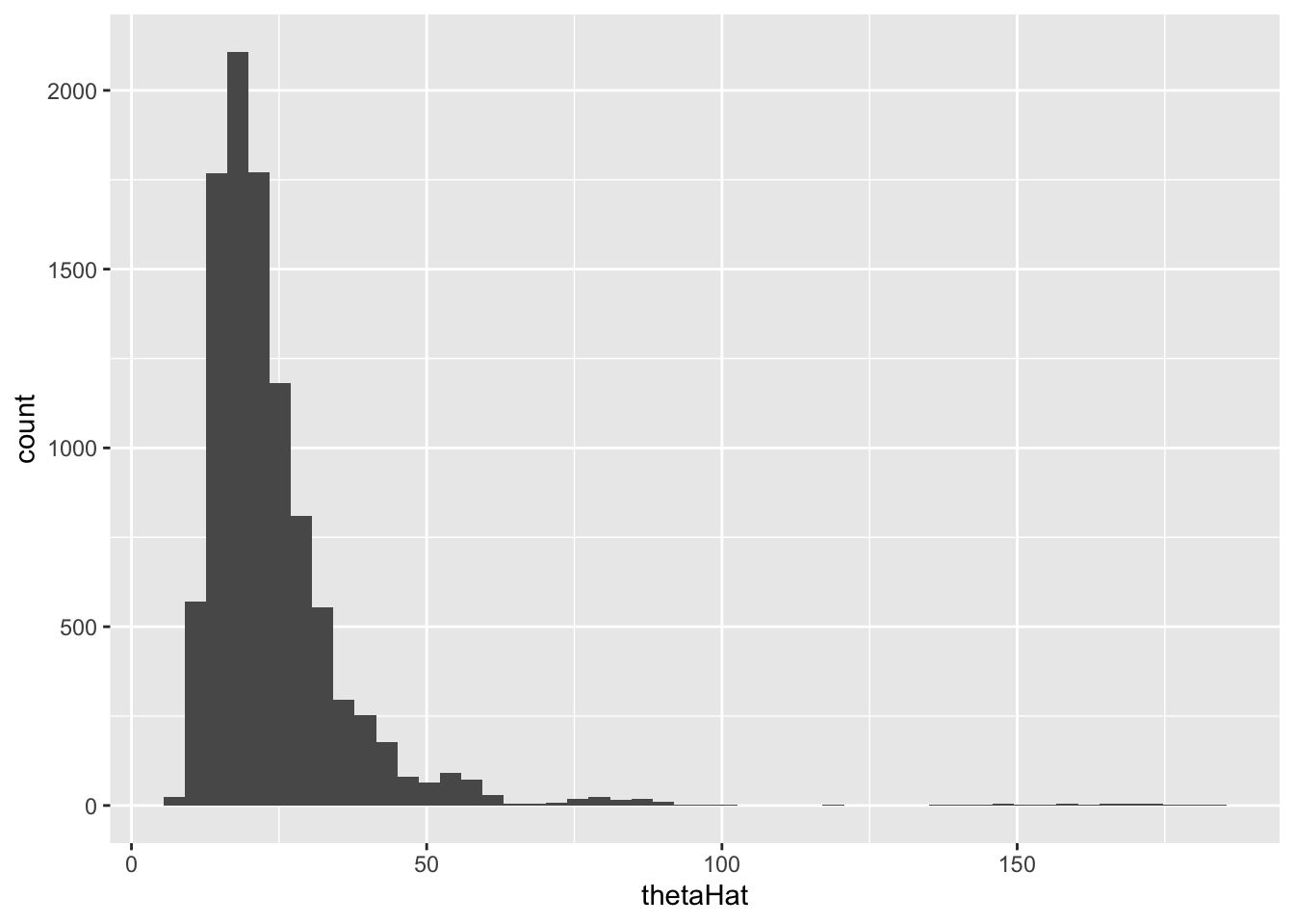
Let’s get a bootstrap 95% confidence interval.
We can say with rigor, that your chances of getting covid with the vaccine are reduced by between 11.339 and 55.867.
We can also do a one-sided 95% confidence interval.
With 95% confidence, your chance of getting infected if vaccinated with Pfizer drops by a factor of at least 12.317
With 99% confidence, your chance of getting infected if vaccinated with Pfizer drops by a factor of at least 10.317
Vaccine efficacy is the 1- the inverse of the ratio above so it is a percentage metric for how getting the vaccine lowers the risk of getting sick.
ratio_eff <- function(data, sample) {
p1 <- sum(data[sample,]$cases == 1 & data[sample,]$patients == 0) /
sum(data[sample,]$patients == 0)
p2 <- sum(data[sample,]$cases == 1 & data[sample,]$patients == 1) /
sum(data[sample,]$patients == 1)
1 - p2/p1 #infected vaccine /infected placebo
}
set.seed(123)
b_out_eff <- boot::boot(trial, ratio_eff, 10000)[1] 0.9506173 2.5% 97.5%
0.91 0.98 These are clinical trial results under controlled conditions. Health organizations also track vaccine effectiveness which is based on how vaccines actually perform under real-world conditions. See Vaccine efficacy, effectiveness and protection.