7 Classification Models
Categrical Data, Binary Classification, Principal Components Analysis, PCA, k-Means Clustering, Support Vector Machines, SVM, Confusion Matrices
7.1 Introduction
This module teaches some classification methods which are useful for working with response variables that are categorical (factors).
Learning Outcomes
- Explain the purpose of classification models.
- Explain the difference between regression and binary classification.
- Build different classification models: PCA, K-means, SVM,
- Tune Classifiers and calculate Performance Metrics
- Explain False Positives and False Negatives from a confusion matrix.
7.1.1 References
- The R Stats Package. See
help("stats"). - Support Vector Machines with {e107} Package (Myer 2024)
- Another package that does SVM and has data sets is {kernlab} package (Karatzoglou et al. 2024)
7.2 Classification
When your question of interest involves a categorical response, you must use a classification method.
- Regression methods/models won’t work with a categorical response very well as they assume the response is continuous.
- The will just generate predictions with decimal places and that does not work for things like “hair color” or “gender”.
There are many Classification methods for binary classification (when there are only two levels) or more general approaches for response variable with multiple levels (categories) such as Hair Color.
Consider a dataset containing several variables (columns). Some of the variables are designated as the “response variables” (or “outputs”). You’d like to predict the response variables from the “explanatory variables” (or “inputs”). There are also variables that you didn’t (or couldn’t) measure, which are the “latent variables”.
These distinctions between variable roles are a bit arbitrary, and it can be a bit of a problem to determine which variables should play which role, especially because the same variable can play different roles depending on your research question.
Exploring your data (on the training set, like in Lesson 6) is an important part of determining possible variable roles.
“Machine learning” is a term that you’ve probably heard, perhaps surrounded by a bit of mystery. While there are several different kinds of algorithms that are usually considered “learning”, they are essentially just fancier versions of statistical regression.
- Machine learning comes in two varieties: “supervised” and “unsupervised”. In supervised learning, the goal is to predict the response variables from the explanatory ones, given that the training data contains the correct answers, just like traditional regression.
- Sometimes you discover new latent variables in the process. In unsupervised learning, the goal is a little different; you want to determine which variables should play which roles.
As in Lesson 6, careful attention to the train-tuning-test process with the data is critical, because modern supervised learning algorithms are extremely flexible.
- Moreover, since they are quite a bit more sensitive to the data than (say) linear regression, it is very easy to get a misleading performance measurement if you don’t follow the correct process.
7.2.1 SVM Overview
We’ll focus our attention on one supervised learning classification technique, called support vector machines (SVMs).
The goal of an SVM is simple: given a table of numerical variables, assign a boolean (true/false) classification to each observation.
You use the training sample of your data to help the SVM give the correct classification to a set of observations with known classifications.
Once “trained” you can use the SVM to classify other observations that don’t have known classifications.
- For instance, a common task for an SVM might be to determine if a pedestrian is in view of a car’s collision avoidance camera.
- Given a digital image – a big collection of numerical variables – the output of such an SVM would simply be “pedestrian” (true) or “not pedestrian” (false).
A given SVM is a rather elaborate algorithm, whose behavior is determined by quite a few numerical “parameters”.
- The training data are used internally to determine these parameters.
- Thankfully, that process is handled internally by the {e1071} package, so you can just tell an SVM to “train thyself” given some data and it will!
Actually, there are many kinds of SVMs, though they ultimately look about the same to the data scientist from the standpoint of inputs and outputs.
- The {e1071} library provides four different types of SVM, though there are many others that are in wide usage.
- In the machine learning jargon, this means that in addition to the parameters of each SVM, there are “hyperparameters” that select the particular SVM algorithm you want to use.
In our train-tuning-test process, the tuning stage, where we select one algorithm without changing it, can be thought of as the training stage for the hyperparameters.
It sounds fancy, but it’s really pretty easy: try a handful of versions of your algorithm, and then pick the best.
It you get an error, you probably do not have the e1071 library installed, and you’ll need kernlab for our dataset. Here is how to fix that:
7.2.2 The spam data set
The data we’ll be using for this lesson is kind of amusing
It is a single table, called spam, in which each row corresponds to an email message.
- The columns are normalized word frequencies for various words present in the textual content of the messages.
- Finally, there is a
typecolumn that either contains the string “spam” or “nonspam”.
You guessed it! We are going to make an email spam filter!
Now it happens that this particular dataset is rather easy to “cheat” because it includes a few oddities in how it was collected.
- Specifically, this dataset was collected before “spear phishing” was common (where a spammer impersonates a trusted sender whose account has been compromised).
- As a result, words that are characteristic of the data collector’s organization are a dead giveaway that a message is “nonspam”.
- These correspond to two columns:
georgeandnum650, so we’ll deselect these.
7.3 Create Validation Sets
As we described earlier, we need to create a sampling frame and split our data into training, tuning, and test sets.
set.seed(1234)
raw_data_samplingframe <- spam |>
select(-george, -num650) |> # Remove the "cheating" columns...
mutate(snum = sample.int(n(), n()) / n())
training <- raw_data_samplingframe |>
filter(snum < 0.6) |>
select(-snum)
tuning <- raw_data_samplingframe |>
filter(snum >= 0.6, snum < 0.8) |>
select(-snum)
test <- raw_data_samplingframe |>
filter(snum >= 0.8) |>
select(-snum)You can save these off in CSV files if you like.
In a few places we will want just the input variables. It will be frequently useful to have the correct answers in a separate table as well.
Let’s split these off now:
7.4 Training stage:
7.4.1 Principal Components Analysis
Let’s start with a quick visualization! Since all the explanatory variables are numerical (they’re normalized word frequencies) and there are quite few of them, principal components analysis (PCA) is a good way to create just two variables that contain most of the information in the data.
Principal Components Analysis (PCA) is a well-established method for reducing the dimensions of the data.
It uses linear algebra to create linear combinations of a set of \(k\) variables into \(k\) Principal Components where each principal component is independent of the others.
The Principal Components are designed so the First principal component captures as much information as possible from one linear combination, then the second captures the most of out what is left, and so on.
Thus the first two principal components are often used in plots as they capture the bulk of the information in just two variables so we can easily plot them.
- Use the
prcomp()function from the Base R {stats} package.
Now we can plot the messages as a scatterplot, and color by type:
spam_pca$x |>
as_tibble() |>
mutate(type = training_truth) |>
ggplot(aes(PC1, PC2, color = type)) +
geom_point()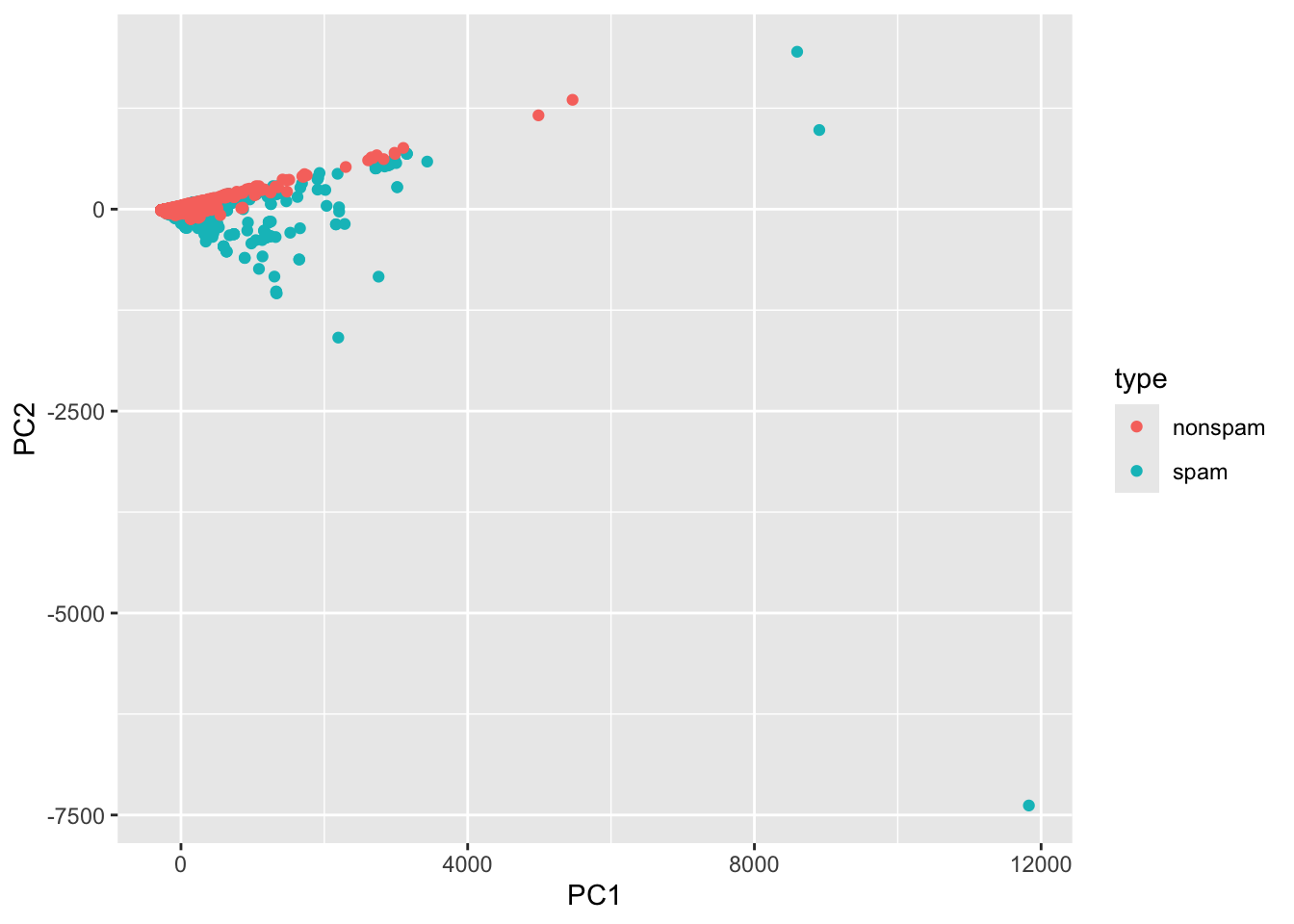
Each message corresponds to a point in this plot.
- Messages with similar word usage end up near each other, while messages using different words tend to be further away from each other.
- Notice how the spam messages spread out a bit further from the nonspam messages.
- These outliers are easier to detect than the spam messages that do a better job of looking like a nonspam message.
7.4.2 K-Means Clustering
Clustering with k-means is a kind of “semi”-supervised learning, in that we tell it the number of classes we want.
K-means clustering is a method for separating data into \(k\) clusters or groups (you pick the \(k\)) such the points in each cluster as as similar to each other as possible and as dissimilar to points in any other cluster as much as possible.
K-means is an iterative algorithm.
- We set the seed as the first step in K-means is to randomly assign all points to one of the possible clusters.
- It then calculates a lot of distances and moves points from one cluster to another.
- It then recalculates the distances and moves points, over and over, until the improvement in performance is minimal.
In the case of spam detection, there are two classes: spam and nonspam. So, here we go!
- Use the
kmeans()function from the Base R {stats} package.
Did k-means correctly identify the spam? We can tell if we make a table comparing the k-means cluster ID (an integer) with the true classes:
Then we can count the number of times we get a match between the two.
We could use table() but I think this looks nicer as a contingency table, so we’ll need to pivot_wider():
table(kmeans_results)
kmeans_results |>
count(training_truth, km) |>
pivot_wider(names_from = km, values_from = n) km
training_truth 1 2
nonspam 1675 31
spam 946 108# A tibble: 2 × 3
training_truth `1` `2`
<fct> <int> <int>
1 nonspam 1675 31
2 spam 946 108Have a look at the results closely.
- The best possible performance consists of having each row and each column having exactly one zero entry.
- Because k-means has an element of randomness to it, I can’t tell you specifically which class ID number (the
kmcolumn) means spam or not, but it’s not very effective.
Just as a sanity check, though, you can ask whether k-means is detecting “something” about the spamminess of a message.
- The way to do this is with chi-squared.
- We’ve done this many times before, so the following block of code should make perfect sense to set up the contingency table:
And then let’s just run the test:
Pearson's Chi-squared test with Yates' continuity correction
data: ct
X-squared = 95.041, df = 1, p-value < 2.2e-16Well, I got a small \(p\)-value. So k-means detects a statistically significant feature of the spam, but it’s not really conclusive either.
Why did k-means do poorly? Let’s label the PCA plot we made in Step 8 with the k-means clusters instead of the true values:
- We use
as.factor()to convert the clusters from numbers to a factor so the legend is correct.
spam_pca$x |>
as_tibble() |>
ggplot(aes(PC1, PC2, color = as.factor(spam_kmeans$cluster))) +
geom_point(alpha = .6) 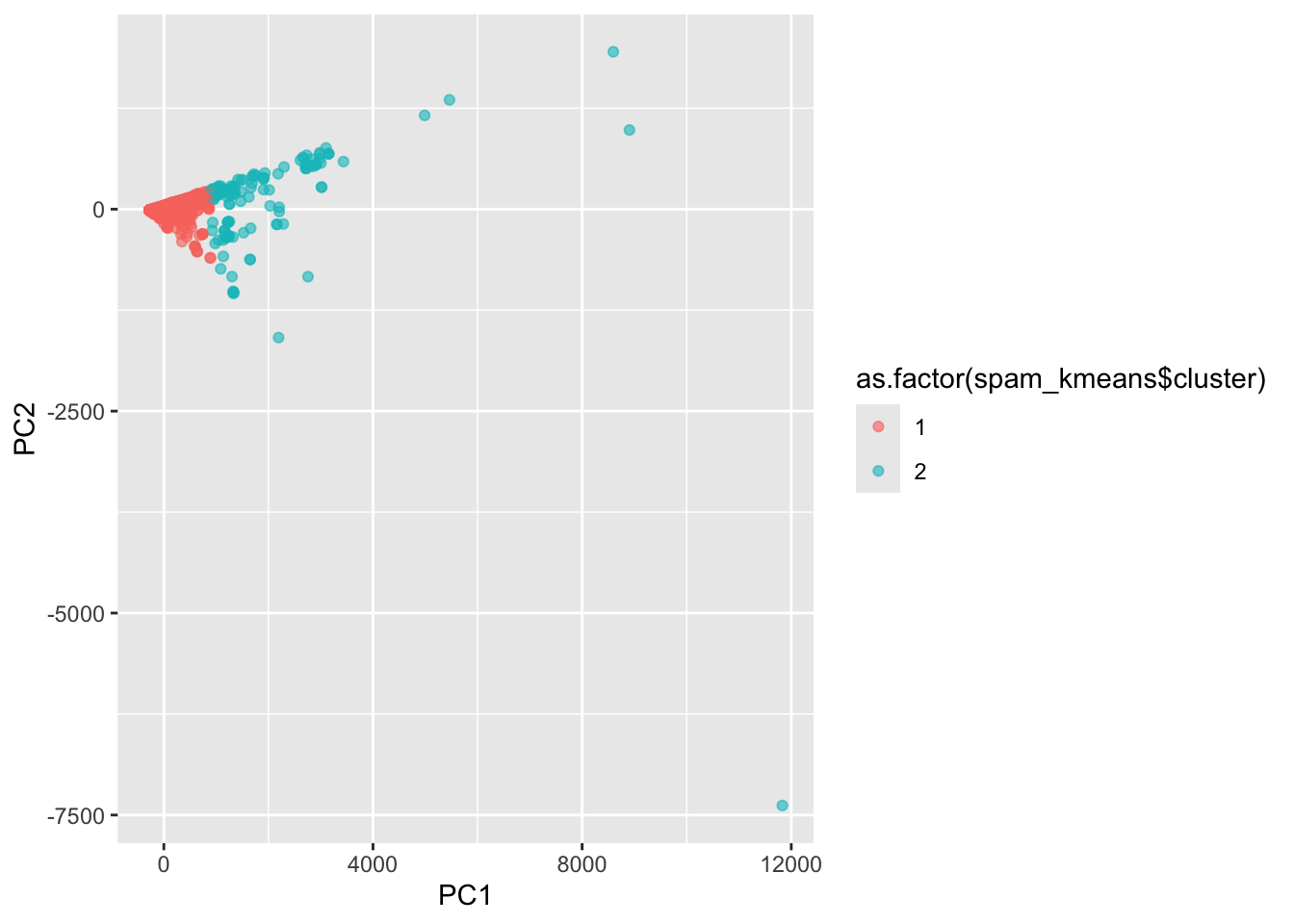
What you should see is that k-means splits the messages into an “inner core” and “outliers”.
- It labels all of the inner core as nonspam, and the outliers as spam.
- While this works for the really far outliers (which are obviously spam messages!), it doesn’t do a good job closer to the core.
Let’s give up on k-means, and switch over to SVM training.
7.4.3 Training the SVM
You train an SVM using the svm() function.
- The
svm()function can take two styles of input, though they both do the same thing internally. If you’re interested, you can read the documentation:
Here’s the format that the e1071 authors use more frequently in the documentation:
- The
~is the formula operator again. - The
.on the right side of the~means use all the variables.
The other option is tidyverse compatible, but you have to split the data into input and output first:
These both do the same thing, so pick whichever you like. I’m going to use the first version in what follows.
The way SVMs work is that they split the space of observations (imagine the PCA plot) into two halves by way of a “separating surface”.
- That surface is high dimensional and takes quite a few parameters to define, but you need not worry too much about them.
- Additionally, there are many kinds of surfaces you might try to use, each defined by a set of equations. - Choose a different set of equations, and your surfaces will look different!
- The choice of a set of equations is called a “hyperparameter”, and in the case of
svm(), the hyperparameter is calledkernel.
There are four kernels supported by e1071:
- ‘linear’ : The separating surface is a flat plane
- ‘polynomial’ : The separating surface is a given by a polynomial… like \(z=ax^2+by^2+cxy+...\) (lots more)
- ‘radial’ : The separating surface is an ellipsoidal blob
- ‘sigmoid’ : The separating surface is kind of “S”-shaped
Since our dataset isn’t too big, it doesn’t hurt to try all of these!
- We can use the tuning dataset to select the one that does the best job at correctly classifying the email messages as spam or nonspam.
How do we do this? Well, we just repeat with a different kernel:
Note: each of the different kernels have additional hyperparameters in them (like degree, gamma, and the like), that you might need to change in some circumstances. - We don’t need to do that here, but you may have to change them if you don’t get good results on other data…
Now that we’ve trained the SVMs for our data, it’s a good idea to anticipate their performance.
7.4.4 Plotting Results
Plotting is perhaps the best way to do this, but plotting in the original data variables is not reasonable.
- That’s unfortunate because that’s how our SVMs were trained.
- But, since we are using the training data, there is nothing wrong with training an additional set of SVMs to work on the PCA data since these plot better.
- The hope (which will be validated against the tuning sample) is that these plots will be representative.
To that end, we need to transform the training data using PCA, and add back the true classifications (‘spam’ / ‘nonspam’).
OK, let’s plot each SVM. It happens that the trained SVMs made by the svm() function already know how to plot themselves, though this doesn’t use ggplot().
- Instead, it uses the base R
plot()function. Here’s how to do it:
sl <- svm(type ~ ., data = training_pca, kernel = "linear") # Watch the punctuation!
plot(sl, training_pca, PC1 ~ PC2, grid = 400) # This plots PC1 versus PC2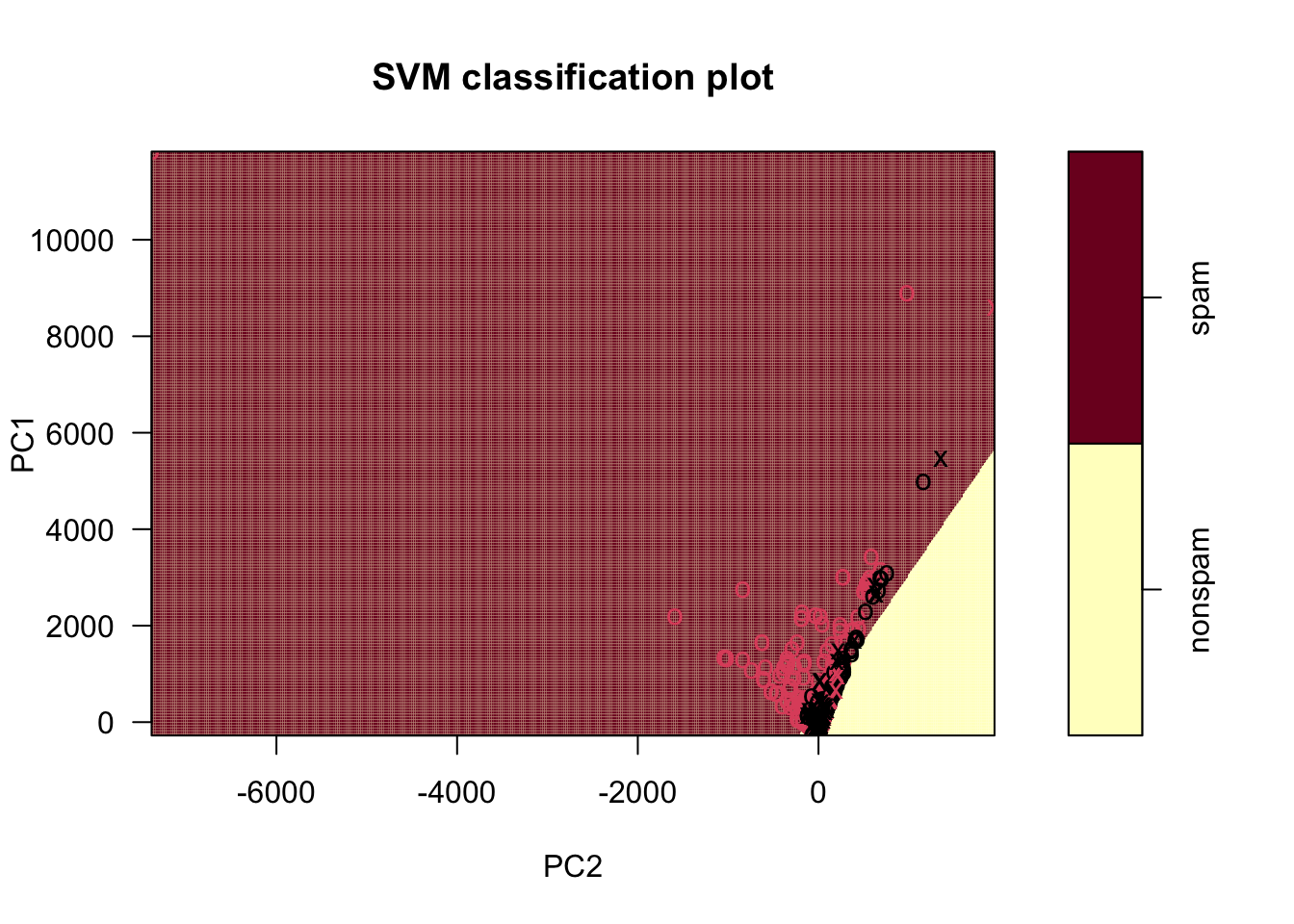
- The points are color coded according to the actual response.
- The
xshapes are the points that are the “support vectors” used to figure out the boundary. - The black points in the red space are misclassified
If you look closely at this, you can see a line slicing through the plot, separating the spam from the nonspam. It does a pretty good job!
Try this with the other three kernels, changing what evidently needs to be changed.
- Notice the different shapes of the different spam/nonspam regions! They all look a little different.
Which do you think best matches the shape of the true classes?
7.5 Tuning stage
Now we’re ready to choose the SVM kernel that will serve as our final version! Since our SVMs are already trained, we can just apply them to the new input data.
The way to do this is via the predict() function. It works like this:
6 8 9 14 20 22 24 28 32 39
spam spam spam spam spam spam spam spam spam spam
45 52 53 55 66 79 88 97 98 101
spam spam spam spam spam spam spam spam spam spam
102 104 106 108 131 135 137 142 145 157
spam spam spam spam spam spam spam spam spam spam
163 165 166 177 188 191 195 201 202 203
spam spam spam spam spam spam spam spam spam spam
211 214 216 222 243 244 249 257 264 265
spam spam spam spam spam spam spam spam spam spam
269 270 290 291 292 293 295 296 307 321
spam spam spam spam spam spam nonspam spam nonspam spam
325 330 331 337 341 342 344 349 367 372
nonspam spam nonspam spam spam spam spam spam spam spam
374 379 380 395 396 404 407 409 415 423
spam spam spam spam spam spam spam spam spam spam
424 425 436 445 447 448 451 452 459 460
spam spam spam nonspam spam spam spam spam spam spam
462 465 467 468 475 479 485 486 489 495
spam nonspam nonspam spam spam nonspam spam spam spam spam
500 502 506 510 512 518 522 526 527 532
spam spam spam spam spam spam spam spam spam spam
534 541 542 561 564 566 567 569 578 579
spam spam spam spam spam spam nonspam spam spam spam
584 586 588 598 615 622 626 630 639 642
spam spam spam spam spam spam spam spam spam spam
644 654 660 665 672 675 679 687 692 694
spam spam spam spam spam spam spam spam spam spam
699 703 707 714 717 721 725 733 734 748
spam spam spam spam spam spam spam spam spam nonspam
751 757 759 766 770 773 778 780 781 785
spam spam spam spam spam spam spam spam spam spam
793 809 810 822 827 831 841 845 851 861
spam spam spam spam spam spam spam spam spam spam
868 869 872 873 874 887 888 894 899 900
spam spam spam spam spam spam spam nonspam spam spam
907 912 914 917 922 926 936 942 943 944
spam spam spam spam spam spam spam spam spam spam
945 946 947 955 957 958 959 961 963 965
spam nonspam nonspam spam spam spam spam spam spam nonspam
968 972 978 979 981 982 984 992 994 1004
spam spam spam spam spam spam spam spam spam spam
1009 1013 1016 1017 1020 1030 1033 1037 1040 1055
spam spam spam spam nonspam spam spam spam spam spam
1071 1075 1077 1079 1082 1087 1092 1096 1099 1102
spam spam spam spam spam spam spam spam spam spam
1103 1106 1116 1117 1121 1126 1128 1131 1133 1136
spam spam spam spam spam spam spam spam spam spam
1139 1149 1150 1160 1170 1174 1186 1192 1193 1194
spam spam spam spam spam spam nonspam spam nonspam spam
1201 1203 1204 1207 1216 1222 1224 1233 1236 1239
spam spam spam nonspam spam spam nonspam nonspam nonspam spam
1240 1250 1253 1254 1258 1259 1262 1268 1276 1283
spam spam spam spam spam spam spam spam spam spam
1286 1290 1292 1293 1297 1299 1310 1314 1318 1320
spam spam spam nonspam spam spam spam spam spam spam
1321 1323 1324 1335 1339 1342 1345 1351 1356 1359
spam spam spam spam spam spam spam spam spam spam
1366 1368 1373 1374 1376 1381 1384 1385 1398 1399
nonspam spam spam spam spam spam spam spam spam spam
1400 1404 1409 1425 1431 1434 1435 1444 1450 1455
spam nonspam spam spam spam spam spam spam spam nonspam
1456 1457 1458 1459 1468 1474 1475 1476 1478 1479
spam nonspam spam spam spam spam spam spam spam nonspam
1484 1487 1489 1491 1492 1499 1513 1517 1518 1524
spam spam nonspam spam spam spam spam spam spam spam
1531 1533 1534 1547 1553 1558 1560 1572 1573 1574
spam spam nonspam spam spam spam spam nonspam spam spam
1577 1583 1586 1598 1602 1603 1610 1628 1630 1631
spam spam nonspam spam spam spam spam spam spam spam
1636 1642 1644 1650 1655 1656 1657 1659 1660 1667
nonspam nonspam spam nonspam nonspam spam spam nonspam spam spam
1668 1672 1686 1693 1694 1695 1702 1707 1708 1718
spam spam spam spam spam spam spam spam spam nonspam
1720 1722 1724 1732 1745 1752 1759 1768 1770 1775
spam nonspam nonspam spam spam nonspam spam spam spam nonspam
1781 1782 1785 1787 1794 1799 1811 1827 1835 1836
spam spam spam spam spam spam spam nonspam nonspam nonspam
1844 1848 1855 1859 1864 1866 1872 1878 1880 1883
spam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam
1887 1896 1902 1915 1922 1953 1960 1962 1973 1974
nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam
1977 1980 1990 1992 1993 1995 1998 1999 2006 2018
nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam
2023 2024 2028 2030 2037 2039 2041 2046 2053 2059
nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam
2060 2061 2069 2072 2078 2080 2081 2083 2089 2095
nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam spam
2099 2110 2114 2116 2118 2120 2131 2136 2137 2144
nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam
2147 2150 2151 2166 2167 2169 2175 2181 2184 2194
nonspam nonspam nonspam nonspam spam nonspam nonspam nonspam nonspam nonspam
2200 2201 2205 2208 2212 2225 2240 2241 2245 2247
nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam
2248 2255 2257 2258 2259 2265 2267 2276 2280 2283
nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam
2284 2286 2289 2290 2293 2304 2306 2318 2321 2335
nonspam nonspam nonspam nonspam nonspam nonspam spam nonspam nonspam nonspam
2336 2344 2347 2360 2361 2370 2372 2373 2384 2399
nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam
2408 2413 2414 2419 2421 2428 2437 2439 2444 2445
nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam
2449 2451 2452 2454 2462 2463 2464 2467 2469 2480
nonspam nonspam nonspam nonspam spam nonspam nonspam nonspam nonspam nonspam
2481 2490 2494 2524 2527 2532 2537 2541 2555 2572
spam nonspam nonspam nonspam nonspam spam spam nonspam nonspam nonspam
2573 2574 2576 2580 2584 2586 2589 2607 2609 2616
nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam spam
2625 2628 2637 2639 2640 2643 2648 2652 2655 2665
nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam
2671 2681 2684 2685 2693 2698 2703 2704 2708 2711
nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam
2716 2738 2751 2753 2761 2763 2765 2768 2770 2777
nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam
2781 2782 2785 2787 2804 2813 2824 2825 2830 2831
nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam
2833 2844 2849 2850 2853 2858 2859 2862 2865 2866
nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam
2873 2875 2878 2881 2887 2893 2894 2903 2906 2907
nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam spam nonspam
2908 2909 2910 2913 2920 2933 2935 2940 2947 2951
nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam
2952 2954 2956 2958 2959 2961 2964 2967 2969 2974
nonspam nonspam nonspam spam nonspam nonspam nonspam nonspam nonspam nonspam
2977 2979 2987 2989 2990 2999 3001 3010 3016 3019
nonspam nonspam nonspam nonspam spam nonspam nonspam nonspam nonspam spam
3021 3023 3028 3039 3040 3042 3052 3053 3061 3069
nonspam nonspam nonspam nonspam nonspam spam nonspam nonspam nonspam nonspam
3072 3074 3078 3087 3093 3097 3102 3108 3109 3118
nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam spam nonspam
3124 3138 3140 3150 3152 3153 3157 3162 3166 3168
nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam spam nonspam
3169 3175 3176 3193 3199 3201 3218 3228 3231 3236
nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam
3237 3245 3254 3259 3260 3263 3265 3269 3271 3276
nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam spam
3279 3283 3287 3288 3295 3299 3305 3306 3308 3309
nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam
3313 3315 3316 3323 3325 3328 3330 3332 3338 3355
spam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam
3360 3366 3384 3387 3389 3399 3403 3408 3409 3414
nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam
3415 3418 3426 3430 3431 3434 3442 3452 3457 3462
nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam spam
3468 3476 3478 3479 3487 3501 3532 3537 3538 3539
nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam
3541 3544 3546 3554 3562 3568 3573 3582 3585 3594
nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam
3595 3609 3610 3614 3618 3623 3625 3629 3639 3651
nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam
3652 3653 3655 3659 3665 3666 3677 3684 3692 3699
nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam
3706 3711 3718 3719 3732 3739 3747 3754 3756 3757
nonspam nonspam nonspam nonspam spam spam nonspam nonspam nonspam nonspam
3758 3762 3764 3776 3779 3794 3803 3809 3821 3822
nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam
3826 3828 3839 3844 3855 3859 3864 3867 3871 3873
nonspam spam nonspam nonspam nonspam nonspam nonspam nonspam nonspam spam
3878 3888 3894 3897 3898 3903 3908 3925 3927 3928
nonspam nonspam nonspam spam nonspam nonspam nonspam nonspam nonspam nonspam
3932 3934 3947 3955 3958 3975 3976 3978 3980 3985
nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam
3991 3992 3993 4002 4004 4006 4011 4014 4027 4032
nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam
4035 4043 4048 4078 4080 4083 4085 4087 4088 4094
nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam
4097 4114 4119 4120 4121 4123 4124 4129 4138 4143
nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam
4148 4152 4154 4160 4167 4169 4178 4190 4194 4195
nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam
4203 4213 4215 4217 4223 4232 4234 4239 4254 4260
nonspam nonspam spam nonspam nonspam nonspam nonspam nonspam nonspam nonspam
4268 4273 4281 4296 4303 4306 4312 4315 4317 4318
nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam
4333 4337 4338 4339 4344 4346 4349 4352 4361 4362
nonspam spam nonspam nonspam nonspam nonspam nonspam nonspam spam nonspam
4367 4369 4377 4379 4380 4387 4399 4403 4406 4408
nonspam nonspam nonspam nonspam nonspam spam nonspam nonspam nonspam nonspam
4410 4411 4418 4428 4432 4435 4447 4456 4458 4460
nonspam nonspam spam spam nonspam nonspam nonspam nonspam nonspam spam
4463 4470 4472 4473 4474 4481 4485 4512 4518 4535
nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam
4541 4564 4573 4579 4583 4586 4587 4588 4591 4601
nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam nonspam
Levels: nonspam spamYou should get spammed with some “spam” and “nonspam”! Not very helpful, but it lets us see that the SVM is working.
Let’s use the tibble() function to bundle it all into one big data frame table:
We want to check how many instances we have of where the tuning_truth matches with each of the other four columns: each one is where we correctly identified a message.
The way that we built the tuning_results table is a little annoying, because we really want to do these comparisons systematically.
So, let’s pivot the data longer by reworking the columns:
After this, we check for matches. There are four kinds of these!
tuning_results2 <- tuning_results1 |>
mutate(
tp = (tuning_truth == "spam" & value == "spam"), # True positives (SVM got it right!)
tn = (tuning_truth == "nonspam" & value == "nonspam"), # True negatives (SVM got it right!)
fp = (tuning_truth == "nonspam" & value == "spam"), # False positive (SVM made a mistake)
fn = (tuning_truth == "spam" & value == "nonspam")
) # False negative (SVM made a mistake)Count up the occurrences of each of these
7.5.1 2x2 Confusion Matrices
A 2x2 confusion matrix shows the performance of a model on a sample of size \(n\) by organizing the \(n\) predictions into each of the four possible outcomes and showing the counts for each outcome.
- A confusion matrix allows one to see how much or how little a model “confuses” the two classifications.
| Actual \ Predicted | Positive (PP) | Negative (PN) |
|---|---|---|
| Positive (P) | True Positive (TP) count | False Negative (FN) count |
| Negative (N) | False Positive (FP) count | True Negative (TN) count |
7.6 Testing stage
With the counts of true/false positives/negatives, there are many possible kinds of scores you can make to determine which SVM kernel works best.
- If you divide the counts by the the number of predictions \(n\) the size of your testing data, you get corresponding “rates”.
Different disciplines tend to prefer some of these scores over others: medical tests often report sensitivity and specificity, while most deep learning researchers prefer F1 scores.
If you’re curious, have a look at
https://en.wikipedia.org/wiki/Sensitivity_and_specificity
Just using the formulas on that page, we can compute a few of these:
tuning_results3 |>
mutate(
accuracy = (tp + tn) / (tp + tn + fp + fn),
sens = tp / (tp + fn),
spec = tn / (tn + fp),
ppv = tp / (tp + fp),
npv = fn / (tn + fn),
f1 = 2 * tp / (2 * tp + fp + fn)
)# A tibble: 4 × 11
name tp tn fp fn accuracy sens spec ppv npv f1
<chr> <int> <int> <int> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 linear 346 502 31 41 0.922 0.894 0.942 0.918 0.0755 0.906
2 poly 184 520 13 203 0.765 0.475 0.976 0.934 0.281 0.630
3 radial 346 511 22 41 0.932 0.894 0.959 0.940 0.0743 0.917
4 sigmoid 335 484 49 52 0.890 0.866 0.908 0.872 0.0970 0.869I noticed that ‘linear’ was the best in my case at least in terms of the accuracy and the F1 score, but it’s awfully close to the ‘radial’ score. This is now a situation where plotting can help, if you like.
Finally, you can repeat with just the svm_linear on the test data!
7.6.1 Another Example
The graphics in Figure 7.1 are from an SVM trained on data about home sales.
We want to classify Home QUALITY a factor with three different levels: LOW, MEDIUM, and HIGH.
Our predictors are
SALES PRICEandYEAR_BUILT.Figure 7.1 (a) show the points overlap so we cannot draw a simple line separating the points.
Figure 7.1 (b) shows the linear kernel.
Figure 7.1 (c) shows the radial kernel.
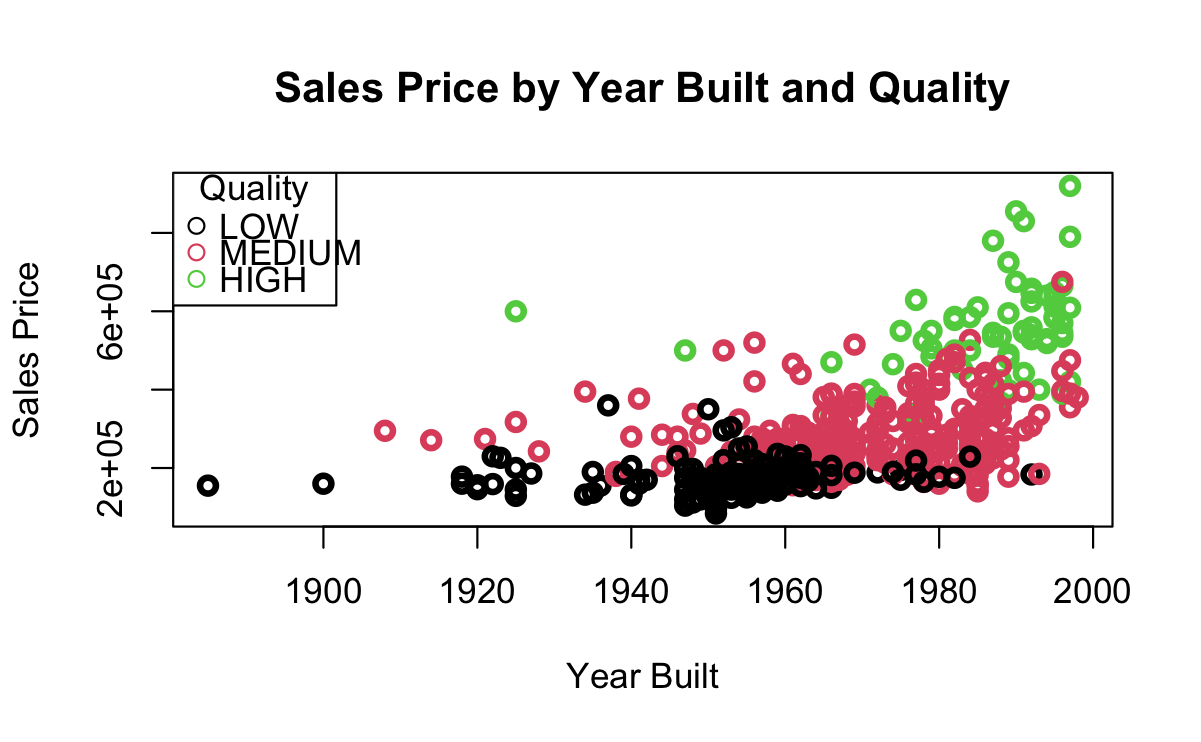
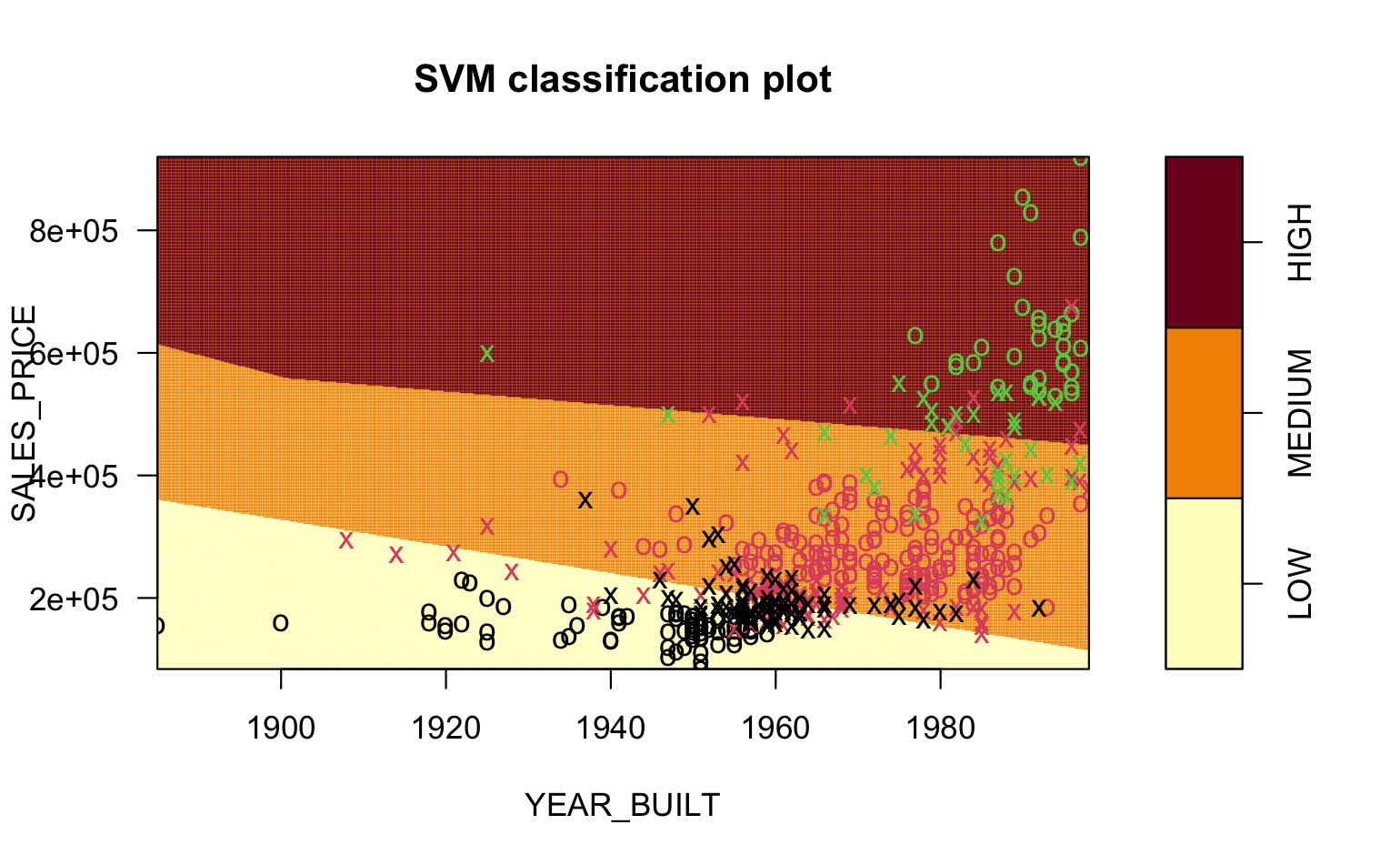
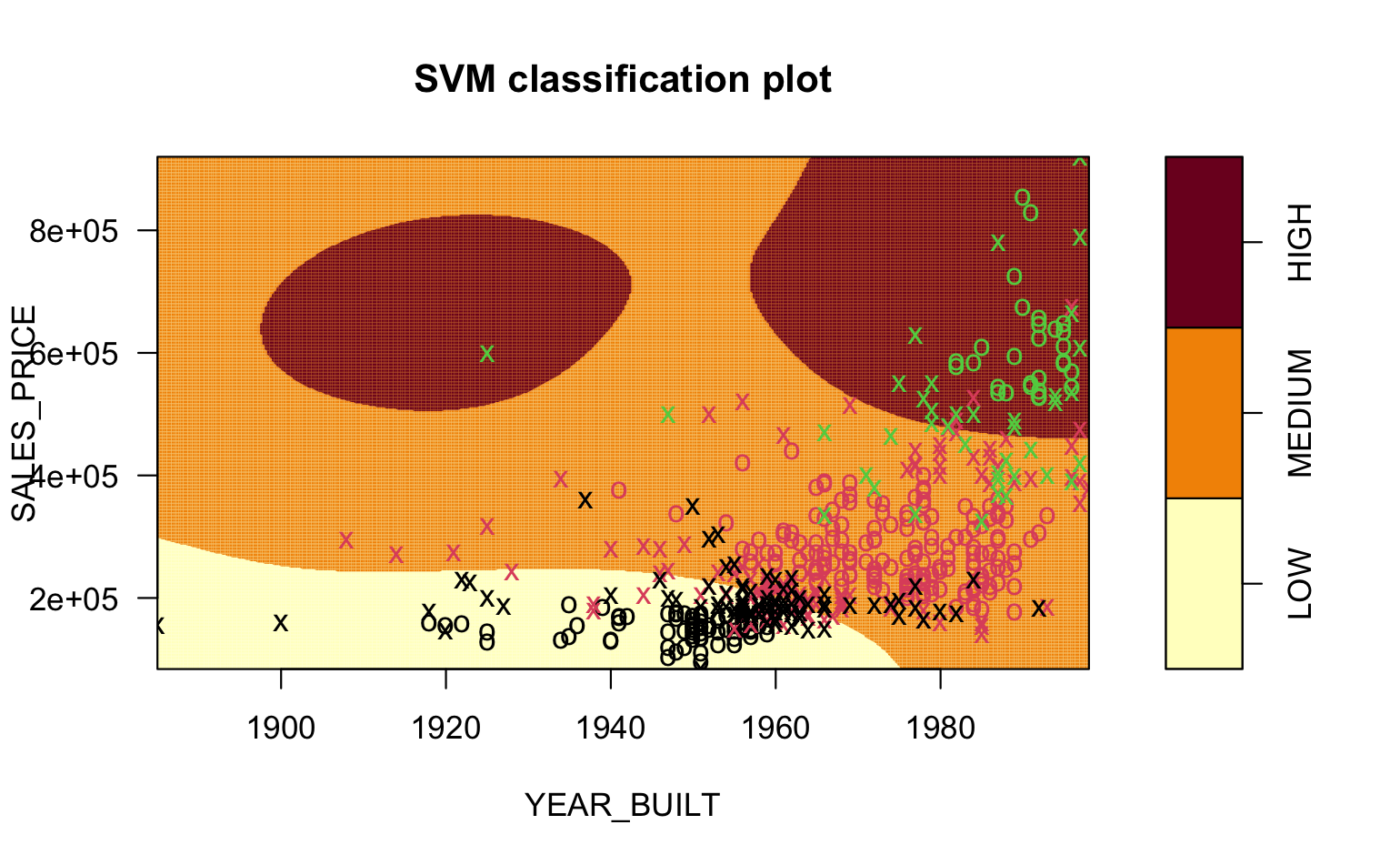
You can see the vastly different shapes of the boundaries for the different regions in the pictures.
- You can see the points that are Errors in prediction, e.g., green circles in the MEDIUM region, red circles in either HIGH or LOW, and black circles in the MEDIUM region.
- Visually, it looks like the Radial kernel might be a better solution.
It can be challenging to visualize what is happening with the different kernels.
In short, a kernel transforms the data into a higher dimensional space, then fits the best multi-dimensional hyperplane it can, and then projects the result back into the original multi-dimensional space.
This YouTube video provides a nice example for a polynomial kernel.
7.7 Summary
Classifiers are an important set of methods for working with categorical response variables.
We have examined just a few forms of classifiers as there are many and there are also many packages with different implementations of the methods.
As with Regression methods, there is no “best classifier” as their performance depends on the underlying (hidden) multi-dimensional structure of the data and how well different methods might fit the data without overfitting.
SVM is a popular method as it supports multiple kernels and is computationally efficient.
7.8 Exercise
- Locate a dataset that has a number of numerical variables that you can use as explanatory variables and a binary (boolean) variable that you can use as a response variable. You could use the dataset from the previous lessons if you like.
Caution: some datasets do not separate well according to the response variables. It’s better if the data separate, but don’t worry if you can’t find a good dataset. The process will “work”, but it might not work well. Be sure to explain your findings; that’s more important than getting good results!
Split your chosen dataset into the three training/tuning/test samples. Save each off each as a CSV file and upload these.
Train at least three SVMs on your training data, being careful to explain which variables are the response and the explanatory variables. Feel free to experiment with different hyperparameters beyond what was done in the worksheet above. The hyperparameters you need might be quite different!
Using your tuning set, select which SVM performs the best… and then…
… determine its performance using the testing set.
Make sure to explain Steps 4 and 5 in comments in your Quarto file!