5 Statistical Tests
t-tests, Chi-squared test, Analysis of Variance, ANOVA, Linear Regression
5.1 Introduction
This module focuses on the use of statistical tests in analyzing data.
Learning Outcomes
- Explain the purpose for statistical tests and a Null hypothesis
- Use and interpret the t.test, aov/anova, and lm functions in R
- Generate residual plots in R.
5.1.1 References
- The R Stats Package. See
help("stats").
5.2 Statistical Tests
Let’s explore the penguins dataset from the previous lessons.
5.3 Subsets and recoding
You can make several sets of data, split on a categorical variable, or recode variables, or create new variables. We already saw an example of this before
As we saw then, recoding can be done in base R or tidyverse, so it’s good to get a sense for how to do the task in both dialects. Let’s use base R this time. Note the “stray” commas below! We are filtering rows, taking ALL columns!
The tidyverse subsetting is a little different. Maybe you’ll like it more… (no tricky commas!)
Although I usually prefer tidyverse, it’s hard to do a clean recoding using tidyverse. Fortunately, whichever way you prefer to filter, it will work.
Now that we have large and small penguins split this way, a t-test is easy!
Welch Two Sample t-test
data: large$flipper_length_mm and small$flipper_length_mm
t = 23.851, df = 160.86, p-value < 2.2e-16
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
22.49402 26.55528
sample estimates:
mean of x mean of y
221.0656 196.5409 If we get a \(p\)-value smaller than 0.05, we can conclude we have found a statistically significant difference between the two groups, at a 95% confidence level!
5.4 Chi-squared Test (Categorical vs. Categorical)
This last graphic we just created begs the question of whether the differences we are seeing in the proportions in the bars is just statistical noise.
Fortunately, we have a test for just this purpose! It’s the chi-square test, and this is how we run it.
newtable <- table(mypenguins$big_penguin, mypenguins$species)
newtable # Shows you what table just did, if you wish to see it.
Adelie Chinstrap Gentoo
FALSE 151 68 62
TRUE 0 0 61chsq <- chisq.test(newtable) # chsq is just a variable to hold your results.
chsq # This gives us a p-value!
Pearson's Chi-squared test
data: newtable
X-squared = 132.19, df = 2, p-value < 2.2e-16
Adelie Chinstrap Gentoo
FALSE 151 68 62
TRUE 0 0 61
Adelie Chinstrap Gentoo
FALSE 124.06725 55.87135 101.0614
TRUE 26.93275 12.12865 21.9386
Adelie Chinstrap Gentoo
FALSE 124.07 55.87 101.06
TRUE 26.93 12.13 21.94If you look at the output from the chisq.test() function, you will see a \(p\)-value. If that \(p\)-value is less than 0.05, we can conclude the differences we see in the ratios in the plot above are statistically significant. Otherwise, we can guess that the differences we are seeing are likely just due to sampling issues and are probably not relevant.
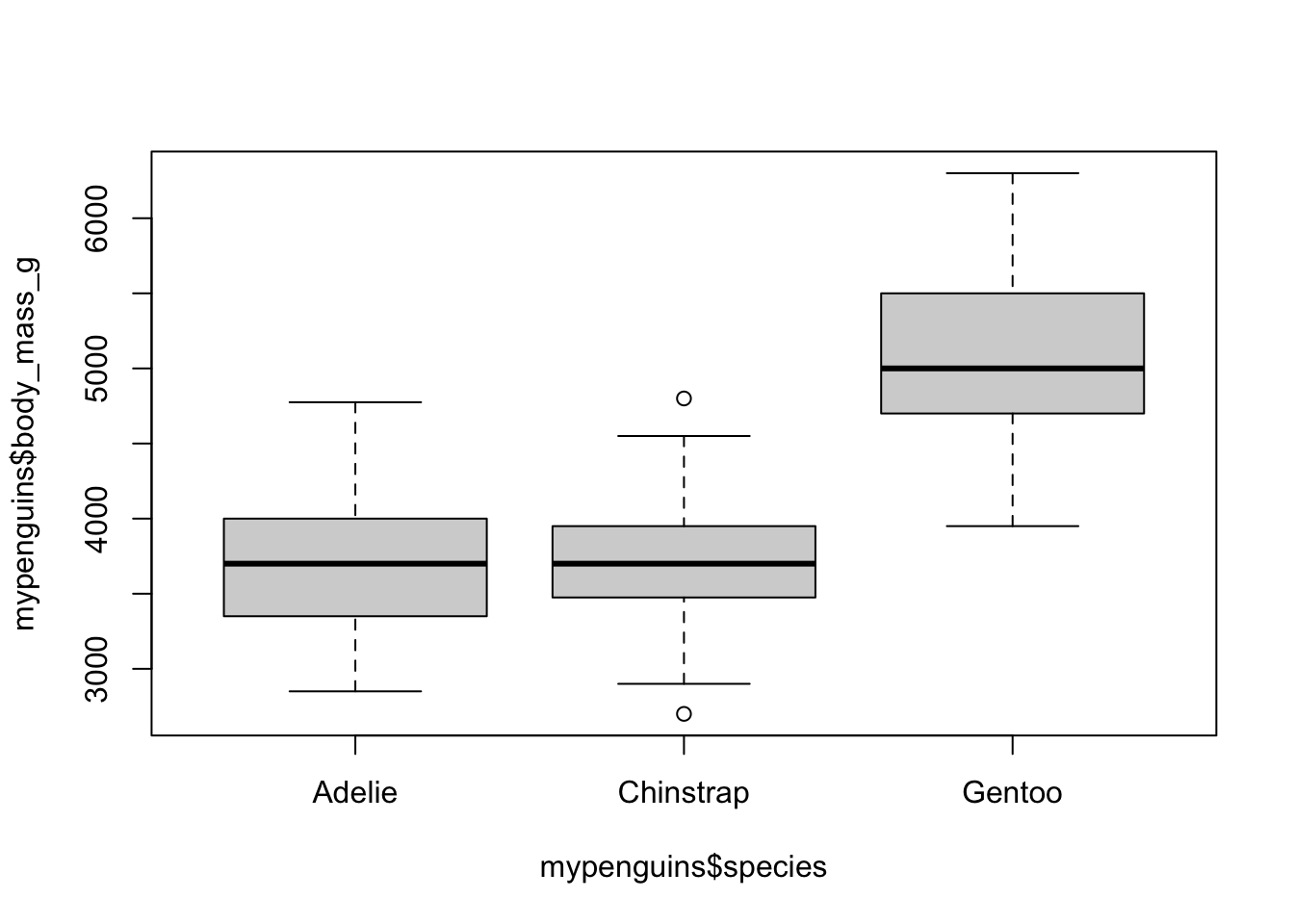
5.5 Box and whisker plots, and ANOVA (Numerical vs. Categorical)
Without using tidyverse, we can get basic boxplots as shown here:
body_mass_g is the numerical variable. species is categorical.
Within tidyverse, as before, we have more options and a nicer graphical presentation of the data.
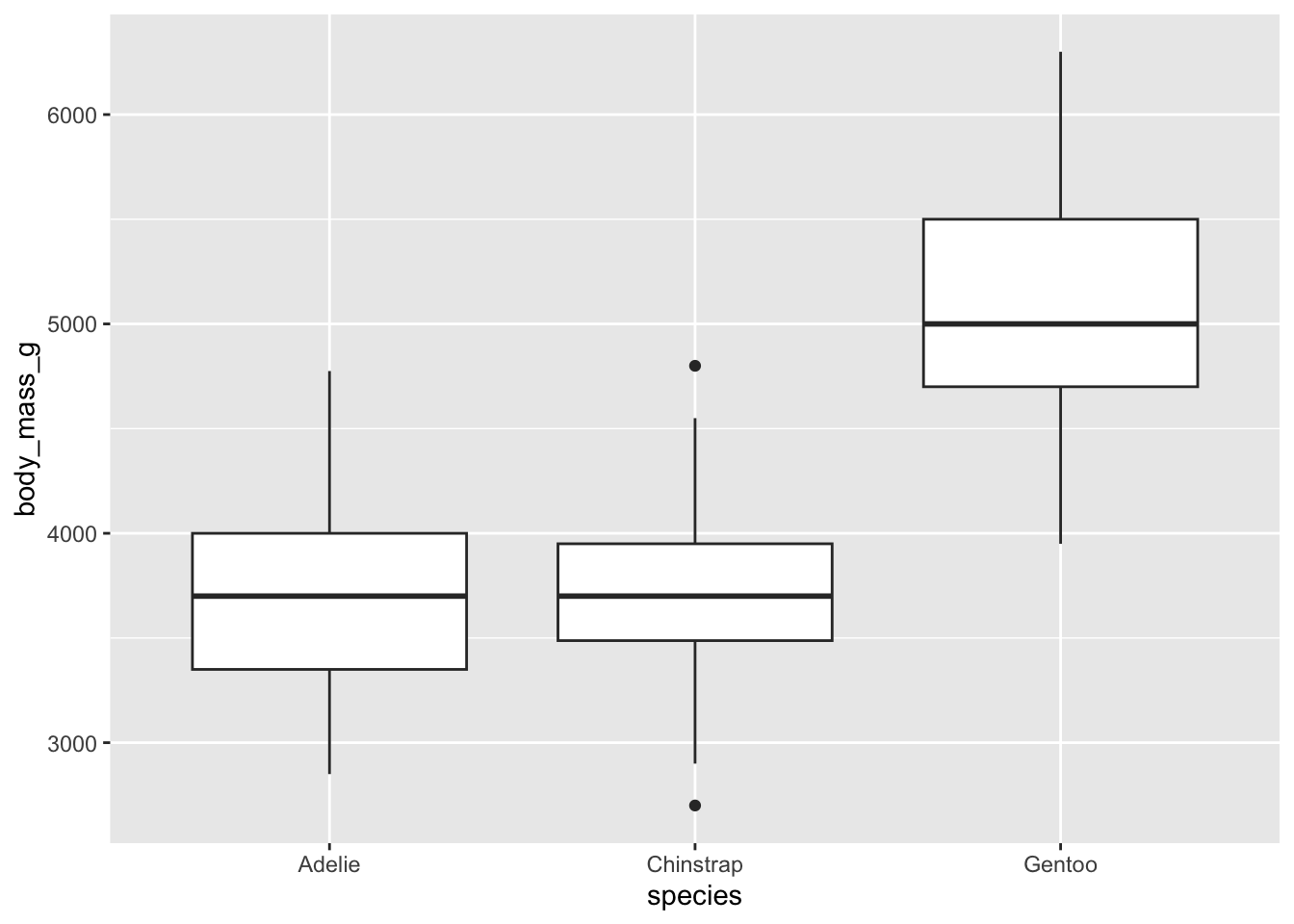
Box plots provide a clear summary of a dataset’s distribution. From a box plot, we can observe:
- The median (central line inside the box),
- The first (Q1) and third (Q3) quartiles (the edges of the box),
- The whiskers, which extend to the smallest and largest values within 1.5 times the interquartile range (IQR = Q3 − Q1),
- Outliers, values beyond the whiskers, are plotted individually as points.
This visualization gives us a good sense of:
- Central tendency (via the median),
- Spread/variability (via the IQR and whiskers),
- Skewness (by checking if the box is symmetric or lopsided),
- And even kurtosis, if there are more outliers than expected, it may suggest heavier tails.
We can make a simpler plot of just the median, min and max if that is of interest.
However, these whisker don’t really extend very far. The default length is probably not what you had in mind. We can override these settings like this:
mypenguins |>
ggplot() +
stat_summary(
mapping = aes(x = species, y = body_mass_g),
fun.min = "min",
fun.max = "max",
fun = "median"
)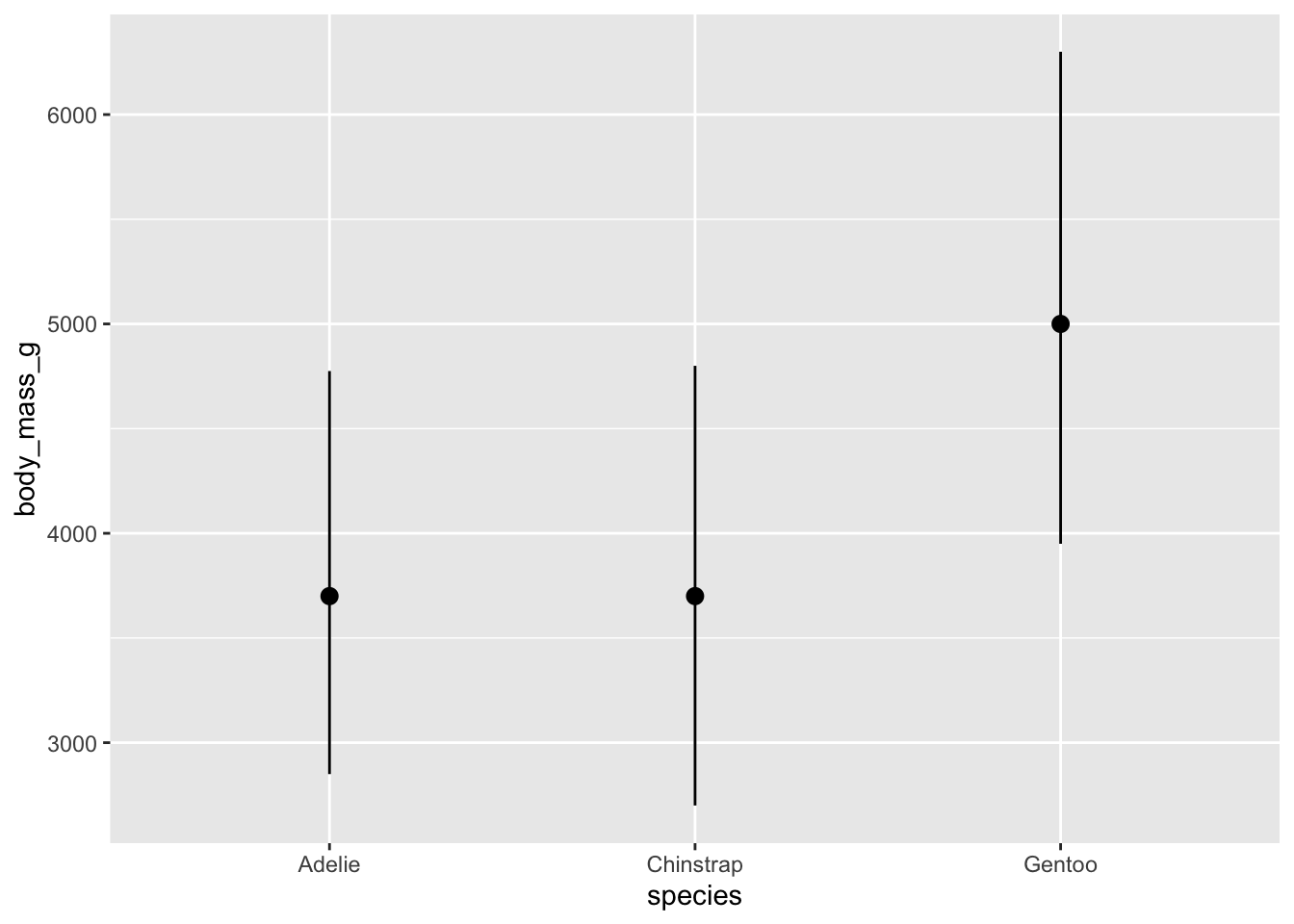
We should see some differences in these bars. One question you ought to be asking is whether or not the differences you are seeing are statistically meaningful. We have a test for this.
It’s either a t-test (if you just have two groups) or ANOVA (for 2 or more groups).
We’ve already used a t-test today. Let’s use ANOVA (ANalysis Of VAriance) this time, because we have three groups.
Df Sum Sq Mean Sq F value Pr(>F)
species 2 146864214 73432107 343.6 <2e-16 ***
Residuals 339 72443483 213698
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
2 observations deleted due to missingnessThe results of the ANOVA should tell us whether the differences we see are meaningful.
- If the p-value is less than 0.05, we can say the differences are significant at a 95% confidence level.
- Here the \(p\)-value is essentially 0 so we would almost never get this F-Value (343.6) if the Null hypothesis, all groups have equal means, were true.
The F-test in ANOVA checks whether the means of multiple groups are equal.
- Null Hypothesis (\(H_0\)): All group means are equal (no difference).
- Alternative Hypothesis (\(H_1\)): At least one group mean is different.
The F-statistic is the ratio of:
\[F = \frac{\text{Variance between groups}}{\text{Variance within groups}} \tag{5.1}\]
If the between-group variance is much larger than within-group variance, the F-value is high, suggesting a significant difference.
The distribution of the F-statistic is not symmetrical and depends upon two parameters, the degrees of freedom for the numerator in Equation 5.1 and the degrees of freedom for the denominator in Equation 5.1.
We can see the df in the results from Listing 5.2 and extract these two values from the aov summary object. Then we can plot.
Show code
summary_result <- summary(aov_result)
# Extract degrees of freedom
df1 <- summary_result[[1]][["Df"]][1] # Between groups
df2 <- summary_result[[1]][["Df"]][2] # Within groups
# Create data frame for F-distribution
# Observed F-value
observed_f <- 343.6
# Critical F-value at alpha = 0.05
critical_value <- qf(0.95, df1, df2)
# Generate F-distribution data
f_data <- tibble(x = seq(0, max(6, observed_f + 1), length.out = 1000)) %>%
mutate(density = df(x, df1, df2))
# Plot
ggplot(f_data, aes(x = x, y = density)) +
geom_line(color = "steelblue", size = 1) +
geom_vline(xintercept = critical_value, color = "red", linetype = "dashed") +
geom_vline(xintercept = observed_f, color = "darkgreen", linetype = "dotted") +
annotate("text", x = critical_value + 0.5, y = max(f_data$density) * 0.9,
label = "Critical value (α = 0.05)", color = "red", hjust = 0) +
annotate("text", x = observed_f - 5, y = max(f_data$density) * 0.5,
label = paste("Observed F =", observed_f), color = "darkgreen", hjust = 1) +
labs(
title = paste("F-distribution (df1 =", df1, ", df2 =", df2, ")"),
x = "F value (log scale)", y = "Density"
) +
theme_minimal() +
scale_x_log10()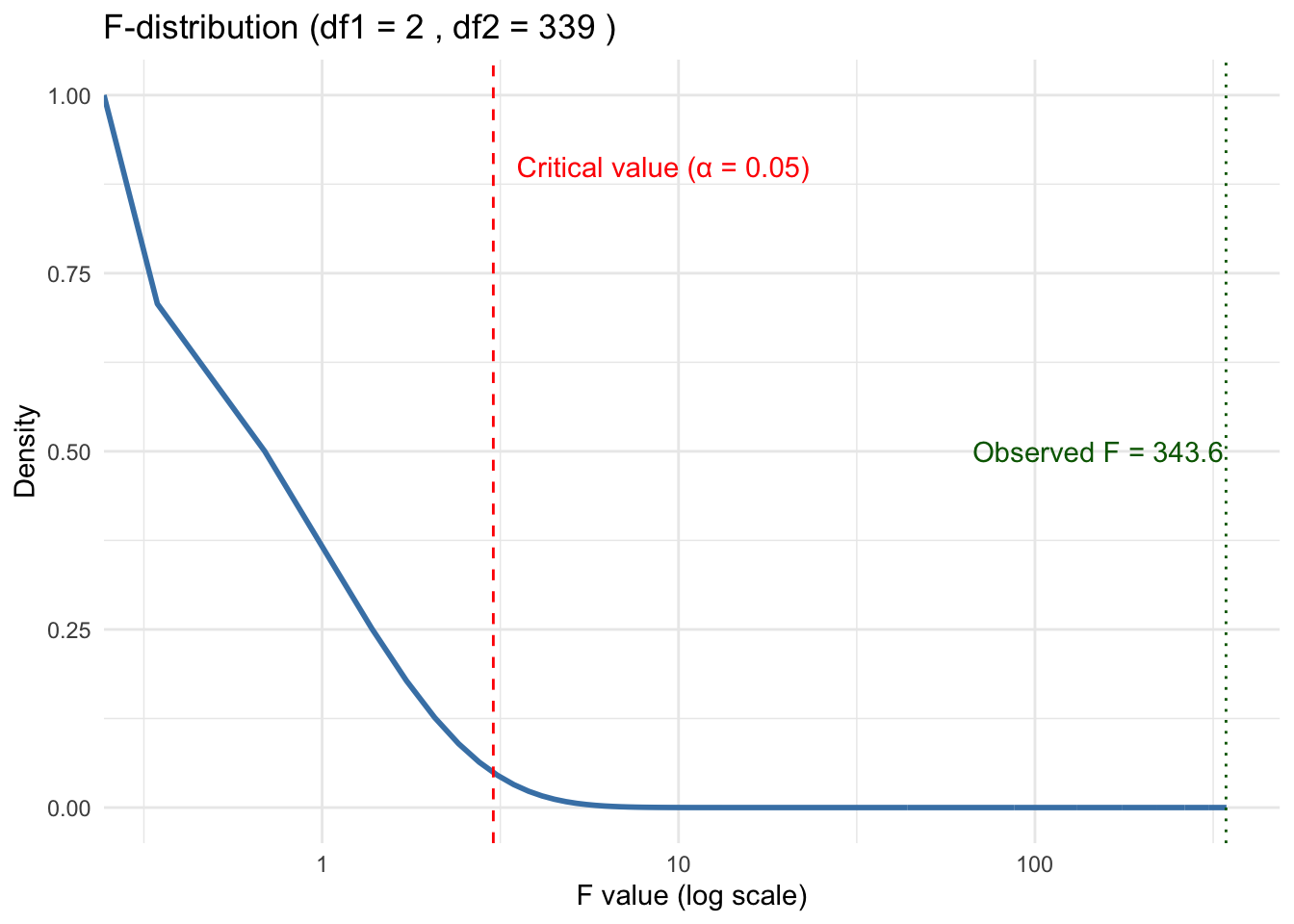
If this happens, we then can move on to Tukey (or Bonferroni) to sort out which groups are actually different from each other.
- It’s pretty clear from the box plot Listing 5.1 which one of these things is not like the other.
- However, we can do another statistical test, Tukey’s HSD (Honestly Significant Difference) test to confirm.
- This is a post-hoc test, performed after, a statistically significant ANOVA result (say, \(p\) < 0.05).
- It compares all pairs of group means and tells you which pairs differ significantly.
Tukey multiple comparisons of means
95% family-wise confidence level
Fit: aov(formula = body_mass_g ~ species, data = mypenguins)
$species
diff lwr upr p adj
Chinstrap-Adelie 32.42598 -126.5002 191.3522 0.8806666
Gentoo-Adelie 1375.35401 1243.1786 1507.5294 0.0000000
Gentoo-Chinstrap 1342.92802 1178.4810 1507.3750 0.0000000- These results show no difference between Chinstrap and Adelie and a highly significant difference, \(p\)-value of 0, between Gentoo and the other two species.
5.6 Linear Regression Tests
Let’s revisit the plot of university and general_revenue_per_person to see if there is a relationship between them and if we can use university to “predict” the general_revenue_per_person.
house_ed_gdp_joined <- read_csv("data/house_ed_gdp_joined.csv")
mywk <- house_ed_gdp_joined |>
mutate(largeFam = Average_size > 3.5)
mywk |>
ggplot(aes(x = university, y = general_revenue_per_person)) +
geom_point() +
geom_smooth(se = FALSE)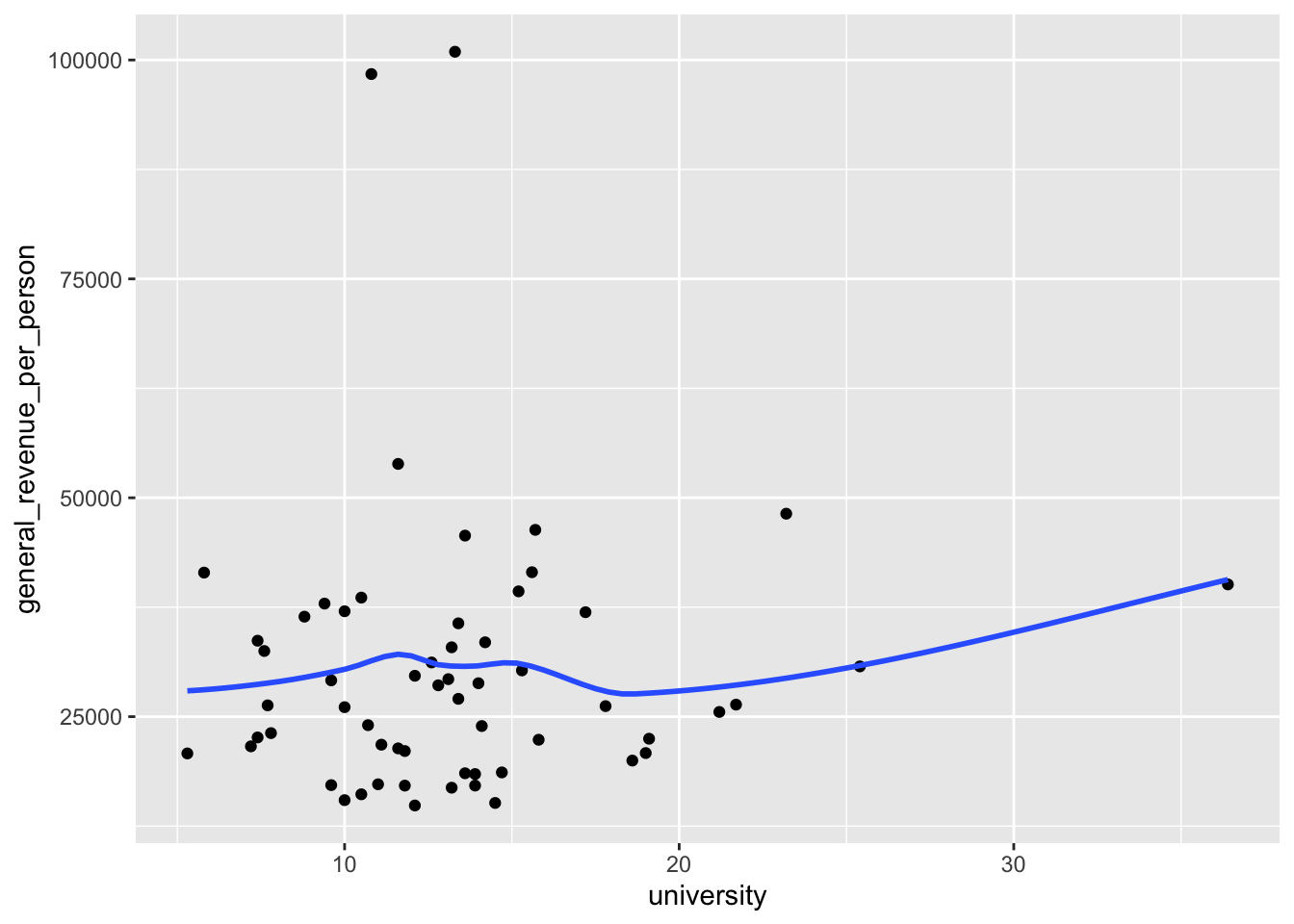
If we convert the smoother to a linear smoother, we see the straight line for a regression model.
mywk |>
ggplot(aes(x = university, y = general_revenue_per_person)) +
geom_point() +
geom_smooth(method = lm, se = FALSE)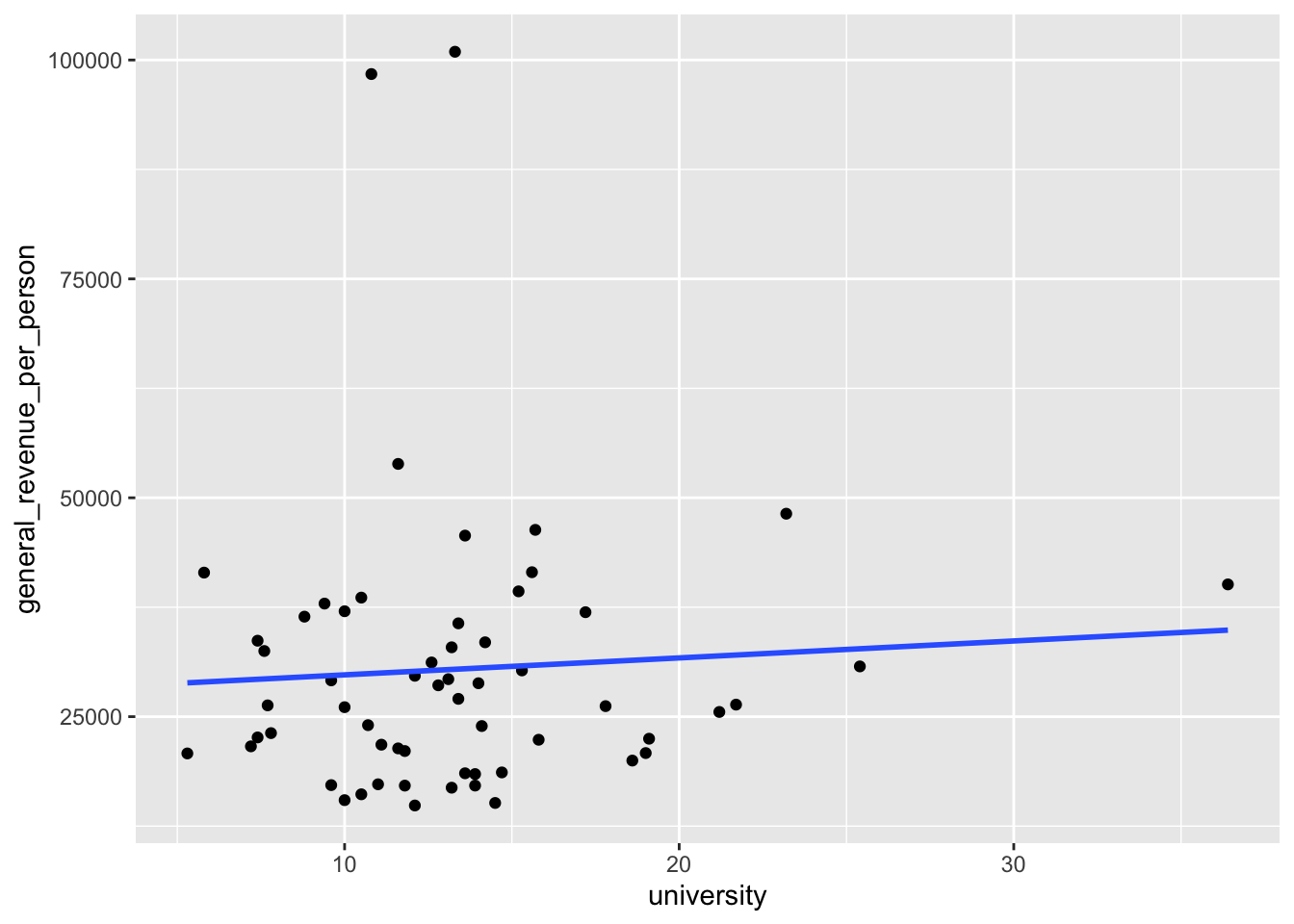
- This regression line suggests there could be a positive relationship between the two variables. But is it real?
Let’s run a test!
- Use the
lm()function to create a linear regression model. - We will use the ~ tilde operator (it’s the squiggly line, not a minus sign) to separate the two sides of our model “formula”.
- The left side is the response we are trying to predict and the right side is all of our predictor variables.
The resulting object has class lm which means is is a linear regression model object.
- It has lots of information about the model.
Show the object and we get the basic results.
Call:
lm(formula = general_revenue_per_person ~ university, data = mywk)
Coefficients:
(Intercept) university
27840.0 193.6 - The coefficients here are the two parameters of the line we saw in Figure 5.1.
- This is known as the “fitted line” and all the predictions are points on this line.
- The intercept is where it crosses the
yaxis and theuniversitycoefficient is the slope of the line. - We interpret that coefficient as saying for every 1 percent increase in
university, thegeneral_revenue_per_personcan be expected to increase by 193.6.
5.6.1 Interpreting the Summary Information on the Coefficients
We can use summary() to get much more information about the model.
- Note that the
summary()function is a generic function that calls special methods for different class objects. Since our model is classlm, the actual function being called issummary.lm()which is what you look for in the help.
Call:
lm(formula = general_revenue_per_person ~ university, data = mywk)
Residuals:
Min 1Q Median 3Q Max
-15501 -9052 -3398 5224 70528
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 27840.0 5883.7 4.732 1.51e-05 ***
university 193.6 411.5 0.470 0.64
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 16150 on 57 degrees of freedom
(2 observations deleted due to missingness)
Multiple R-squared: 0.003867, Adjusted R-squared: -0.01361
F-statistic: 0.2213 on 1 and 57 DF, p-value: 0.6399The summary tells us several items of interest.
- The formula used to build the model.
- The Residuals statistics about the differences between the observed response values and the predicted values (i.e., the residuals).
- Coefficient estimates, along with additional diagnostic statistics:
Std. Error: The standard error of the estimate — a measure of how much the estimate would vary if you repeated the experiment.t value: The t-statistic, calculated by dividing the estimate by its standard error.Pr(>|t|): The probability of observing a t-value as extreme as the one calculated, assuming the Null Hypothesis is true. Smaller values indicate stronger evidence against the Null Hypothesis.- For example, a value of 0.05 means there’s a 5% chance of observing such a result under the Null — considered rare enough to reject it.
- A large value like 0.66 suggests the result is common under the Null, providing little reason to reject it.
Signif. codes: A legend showing how significant a result is using , , or .
What is this Null Hypothesis and why does it matter?
The Null Hypothesis is a statement about the “true” value of a coefficient — in linear regression, it’s typically assumed to be 0. Why?
- If the true value is 0, we can use known statistical distributions to compute the probability of observing a non-zero estimate.
- A statistic that is considered unlikely under the Null is called significant.
In practice, the Null Hypothesis is sometimes jokingly called the “Dull Hypothesis”, because it’s asserting that nothing interesting is happening, i.e., there’s no relationship between the predictor and response variable.
So we now know enough to determine if any of coefficients are significant.
- The intercept has a \(p\)-value that is close to zero so it is rare. That means we can reject the null hypothesis that the intercept = 0.
- The variable
universityhas a \(p\)-value of .64. That is quite high so this variable is Not significant. There is almost no evidence of a relationship between the two variables.
The top part of the summary() output focuses on the individual coefficients. Each variable in the model formula gets a line of output, which lets us determine which variables are statistically significant and which are not.
5.6.2 Interpreting the Summary Information on the Overall Model
Now, let’s look at the bottom portion of the output. This contains information about the overall model:
- Residual standard error: 16150 on 57 degrees of freedom
- This tells us the standard deviation of the residuals (i.e., the errors between predicted and actual values).
- While the number itself isn’t very informative in isolation, it becomes useful when comparing models.
- The degrees of freedom (df) are more interesting. We had 61 total observations, but 2 were deleted due to missing values. Of the remaining 59, 2 were used for the model (one for the intercept, one for the predictor university). So: \(61 - 2_{\text{missing}} - 2_{\text{model}} = 57 \text{df}\)
(2 observations deleted due to missingness) A note explaining why there’s a drop in degrees of freedom.
Multiple R-squared: 0.003867, Adjusted R-squared: -0.01361
- The
Multiple R-squaredmeasures how much variation in the response variable is explained by the model. A value like 0.90 would be considered strong, but here the value is very low, suggesting the model explains almost none of the variation in the data. - The
Adjusted R-squaredadjusts the multiple R-squared based on the number of predictors in the model. This helps prevent overfitting by penalizing the addition of unnecessary variables. If we kept adding variables, \(R^2\) might increase even if those variables are meaningless so using adjusted \(R^2\) helps counteract that.
- F-statistic: 0.2213 on 1 and 57 DF, p-value: 0.6399
- The F-statistic tests whether the entire model is statistically significant. The Null Hypothesis here is that all coefficients (except the intercept) are zero, in other words, none of the predictors have an effect.
- A small \(p\)-value would give us evidence to reject this Null Hypothesis. But here, the \(p\)-value is 0.6399, which is not rare. We have “insufficient evidence” to conclude that the model explains a meaningful portion of the variation in the response.
- Note that since we only had one variable in the model the F-statistic \(p\)-value is the same as the t-statistic \(p\)-value.
So, our tests tell us that despite the slightly positive slope in Figure 5.1, there is no real relationship. That line is just the result of forcing a line on random variation.
5.6.3 Residual Plots
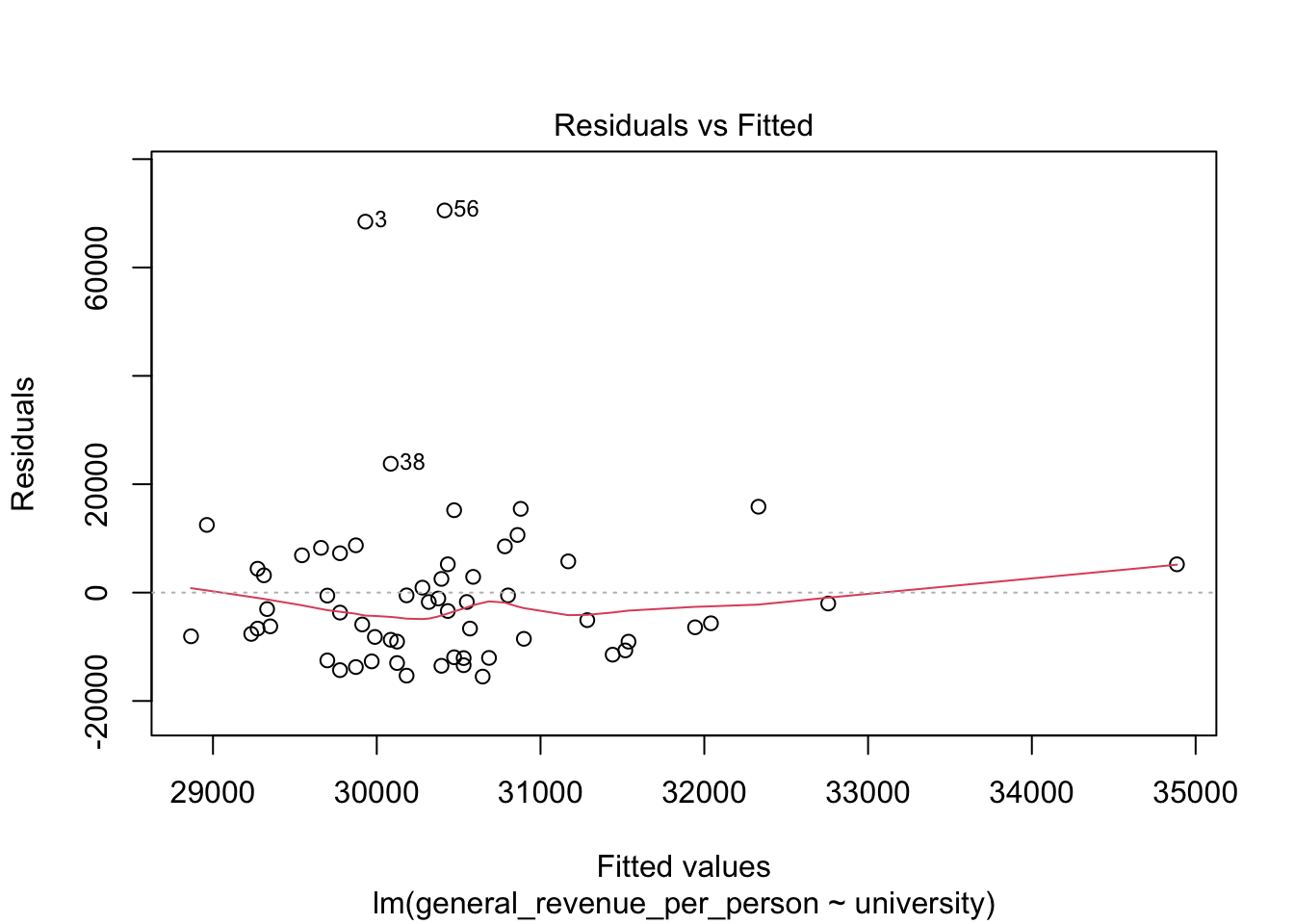
We won’t discuss these now, but the base R plot function has a special method, plot.lm() for doing quick plots for a liner regression model.
- These plots are designed to help you interpret how well the model fits the data and the statistical assumptions about the data that support generating the \(p\)-values.
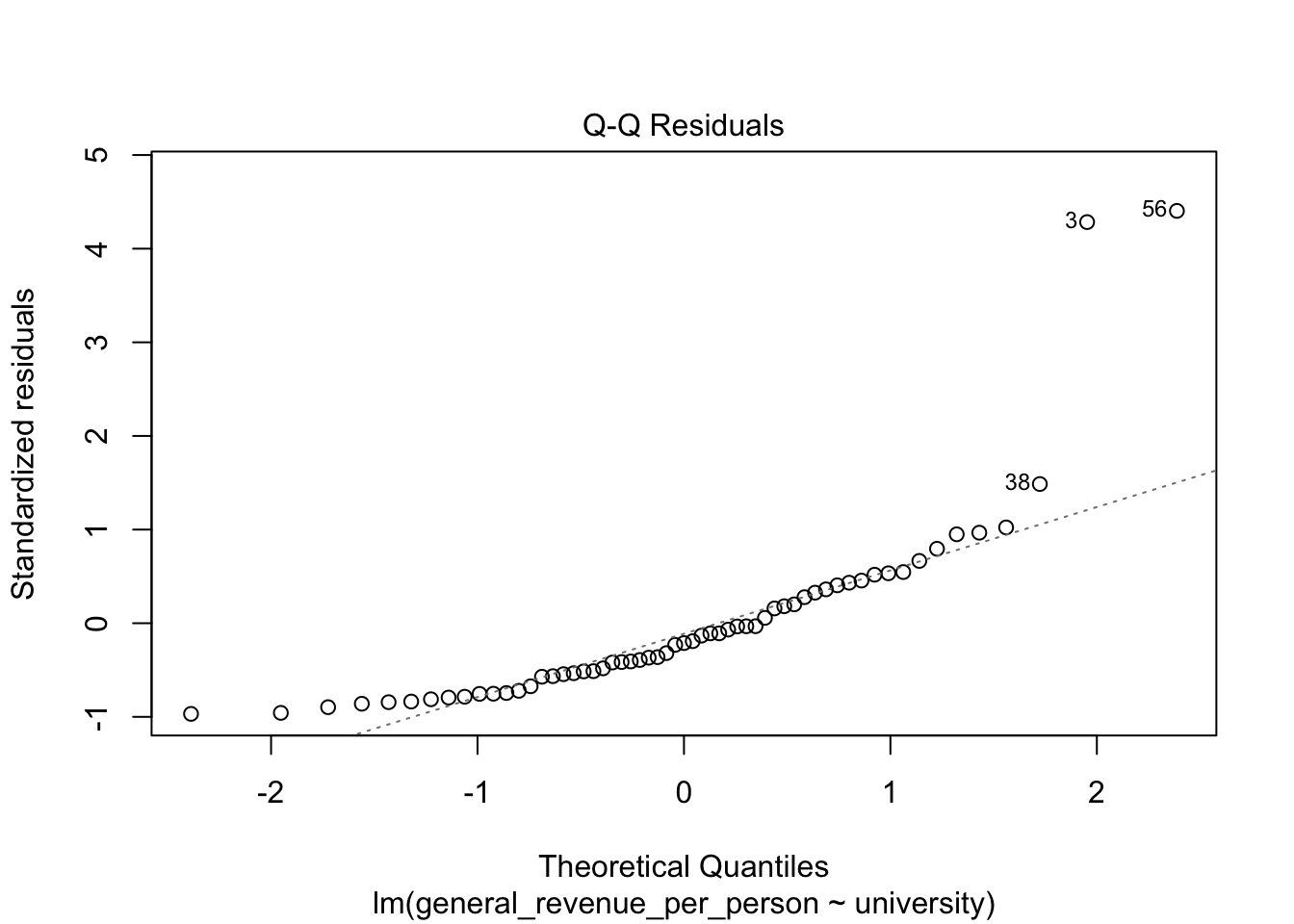
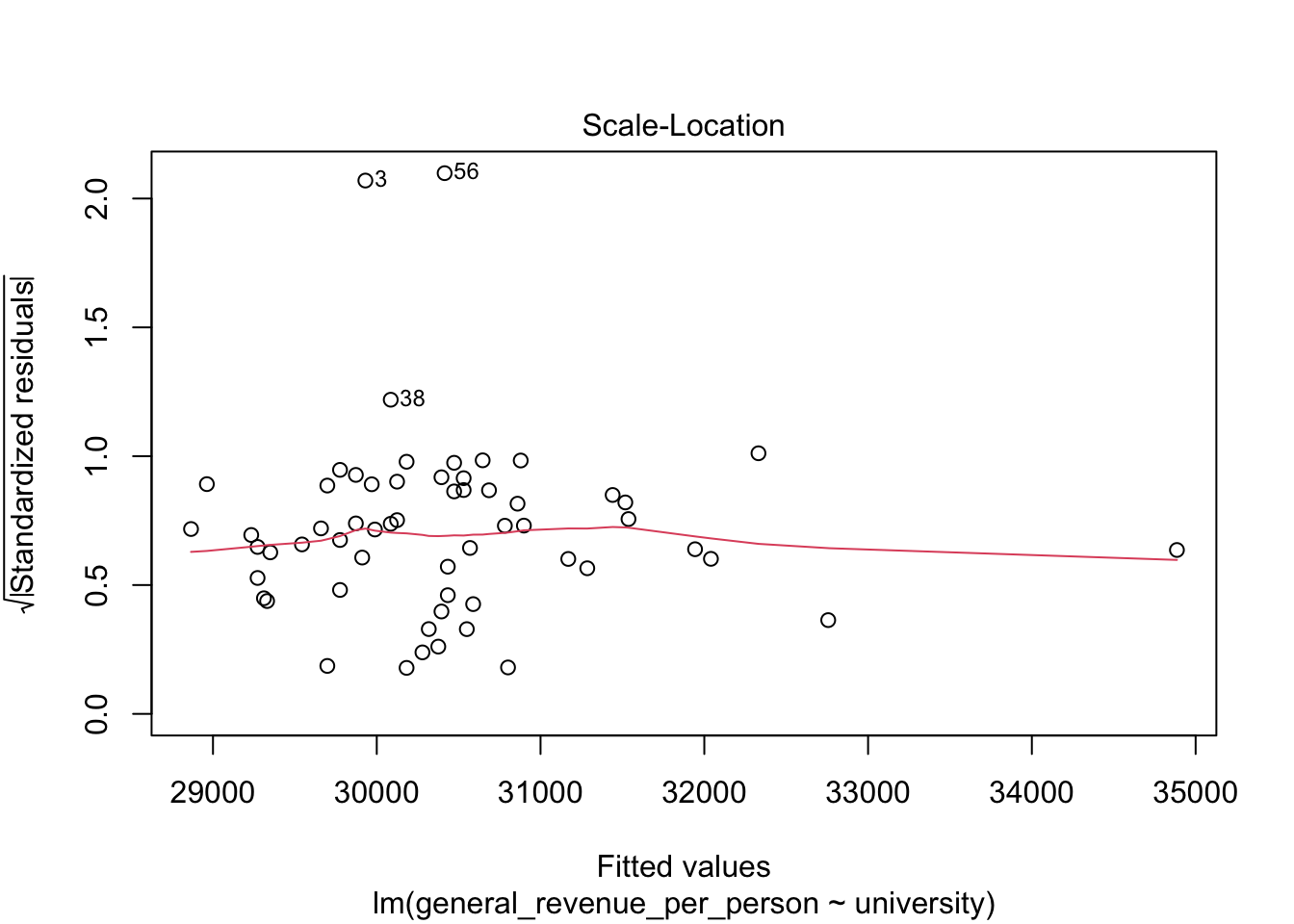
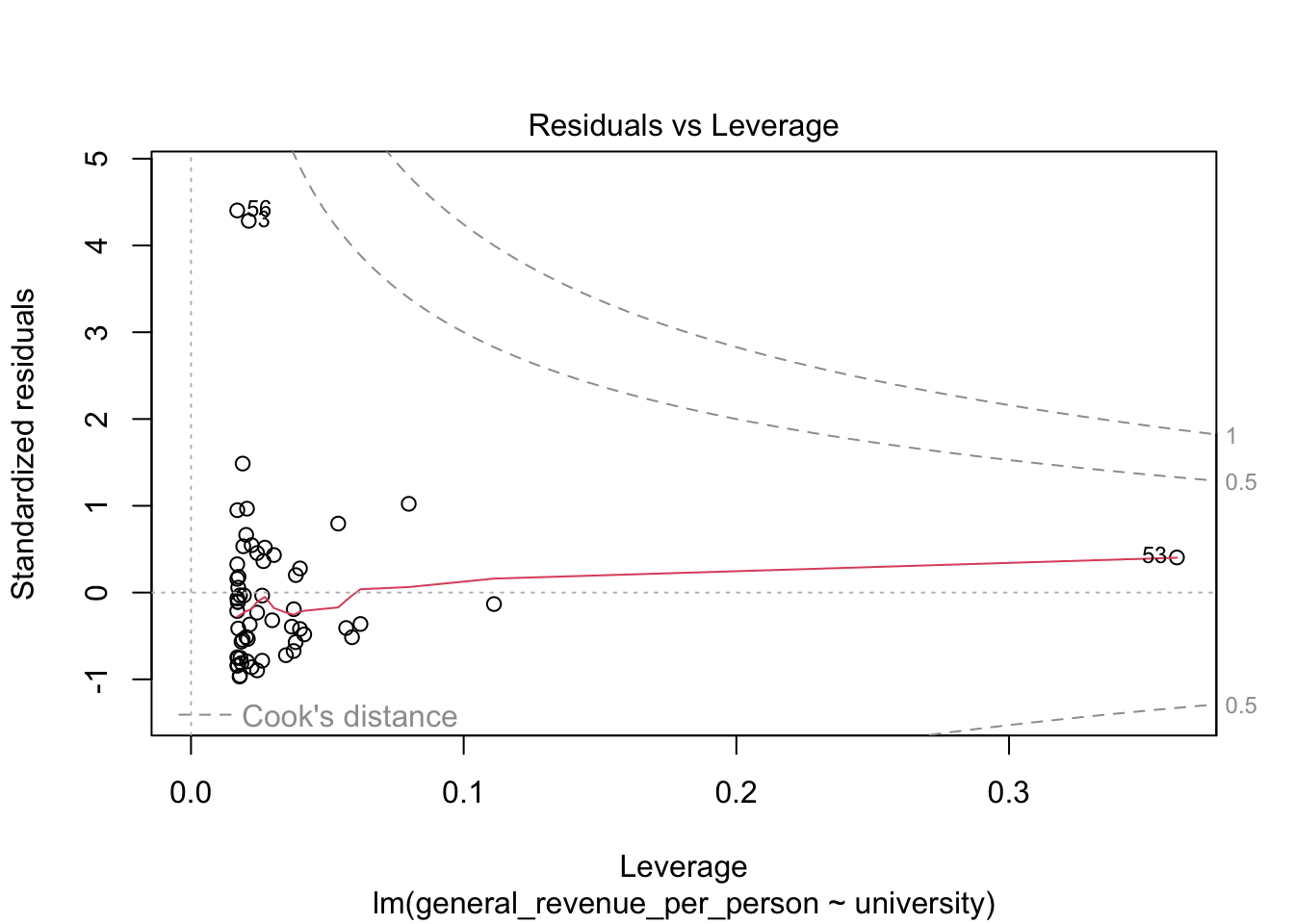
5.6.4 Multivariate Regression Tests
It can be a challenge to plot additional variables in 3, 4, 5 dimensions but it is easy to add them to the model.
Let’s add has_computer and population to see if they help.
my_model2 <- lm(general_revenue_per_person ~ university + has_computer +
population, data = mywk)
summary(my_model2)
Call:
lm(formula = general_revenue_per_person ~ university + has_computer +
population, data = mywk)
Residuals:
Min 1Q Median 3Q Max
-17641 -9919 -3166 5366 67713
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 2.702e+04 7.360e+03 3.671 0.000548 ***
university 1.051e+03 6.709e+02 1.567 0.122904
has_computer -5.718e+02 4.702e+02 -1.216 0.229107
population -1.949e-02 3.915e-02 -0.498 0.620585
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 16070 on 55 degrees of freedom
(2 observations deleted due to missingness)
Multiple R-squared: 0.04914, Adjusted R-squared: -0.002723
F-statistic: 0.9475 on 3 and 55 DF, p-value: 0.4241The summary now shows multiple lines with the additional variables. Unfortunately, the \(p\)-values are all large.
While the Multiple R-squared has increased, the Adjusted R-squared is actually negative which means adding these variables does nothing to improve the model.
So begins the quest of the data scientist to see if they can find variables to explain a quantity of interest!
5.7 Exercise
You can continue with the data from the previous two lessons.
- Subsetting
Demonstrate how to create at least two subsets from your main data, by splitting according to either a Boolean condition, or by a categorical variable. (You are not required to recode a variable.)
Demonstrate a t-test using two subsets you created.
Draw a linear (or smooth) fit
Choose two numerical variables for this. Maybe a linear is not a great choice. Why or why not?
- Demonstrate the use of a Chi-Square test based on two of the categorical variables in your data.
You might be inspired by your previous bar plots.
- Try an ANOVA test!
You might be inspired by your previous box plot.