3 Univariate (Single Variable) Analysis
exploratory data analysis, univariate statistics
3.1 Introduction
This module focuses on the analysis of individual variables - univariate data.
Learning Outcomes
- Generate statistics for variables
- Create univariate plots using the {ggplot2} package
- Customize {ggplot2} plots
3.1.1 References
- Elegant Graphics for Data Analysis {ggplot2} package (Wickham 2016)
- {palmerpenguns} data package (Horst, Hill, and Gorman 2020)
3.2 Univariate Data Analysis
For this demo, a data set from palmerpenguins will be used.
But for your assignment, you need to choose your own dataset! Every student should choose a different dataset for this assignment, which is easy because there are hundreds of freely available datasets! You can use one of the datasets mentioned earlier in the course, or you can use one that comes with an R package (you may need to install the library which contains your data set), or you can get one fresh from the internet.
Let’s also use the dataset we created in the previous lesson.
There is a categorical variables: culture_site, and many numerical variables.
3.3 An initial peek at the data
The R programming language was first released in 1995 but a stable version was not available until 2000. It was inspired by the language S which was created in 1976. Several things have worked in favor of R becoming popular. One is: it is free - like Python, and unlike SPSS, SAS, STATA and S. It is also quite powerful numerically. The other thing that makes R very popular is that it’s incredibly easy to build and distribute new packages for it. Many people who have worked in R have found R to be riddled with annoyances and hassles of various sorts. However, since it’s free and powerful, and since it’s fairly easy to make packages to go along with R, you can probably figure out what happened.
The tidyverse collection of packages arguably contains the most downloaded R packages today. It was introduced to the R community by Hadley Wickham and others starting in about 2016, and we will make heavy use of it for this assignment.
However, you also have the basic R functionality. To give you a feel for both the basic R world and the tidyverse world, you will see similar skills demonstrated both ways. You will probably appreciate tidyverse more after seeing the “low tech” graphics!
Here are a few base R commands to look at the data we’re going to use:
[1] 3.242623[1] 39563.95[1] 257442323[1] 16045.01[1] -0.1281294These can be done using tidyverse as well.
# A tibble: 1 × 1
`mean(Average_size)`
<dbl>
1 3.24# A tibble: 1 × 1
`mean(population, na.rm = TRUE)`
<dbl>
1 39564.# A tibble: 1 × 1
`var(general_revenue_per_person, na.rm = TRUE)`
<dbl>
1 257442323.# A tibble: 1 × 1
`sd(general_revenue_per_person, na.rm = TRUE)`
<dbl>
1 16045.# A tibble: 1 × 1
`cor(Average_size, has_washing_machine)`
<dbl>
1 -0.128For the moment, this seems to be considerably more verbose. Often tidyverse code is easier to understand and maintain, but it really is a matter of preference.
3.4 Counts of categorical variables: Histograms and bar plots
Bar plots (bar charts) are nearly identical to histograms, but bar plots store counts for categorical data, while histograms give counts for continuous numerical data which has been placed into bins for convenience. To a human, this can be a minor difference, but to a computer, it’s a significant difference. So, when talking to a computer (or to a statistician), you must be careful not to confuse the two. Some computer commands are flexible and are designed to take both categorical as well as numerical binning variables as input, but most are not.
Here is the base R command to get a histogram and then a bar plot:
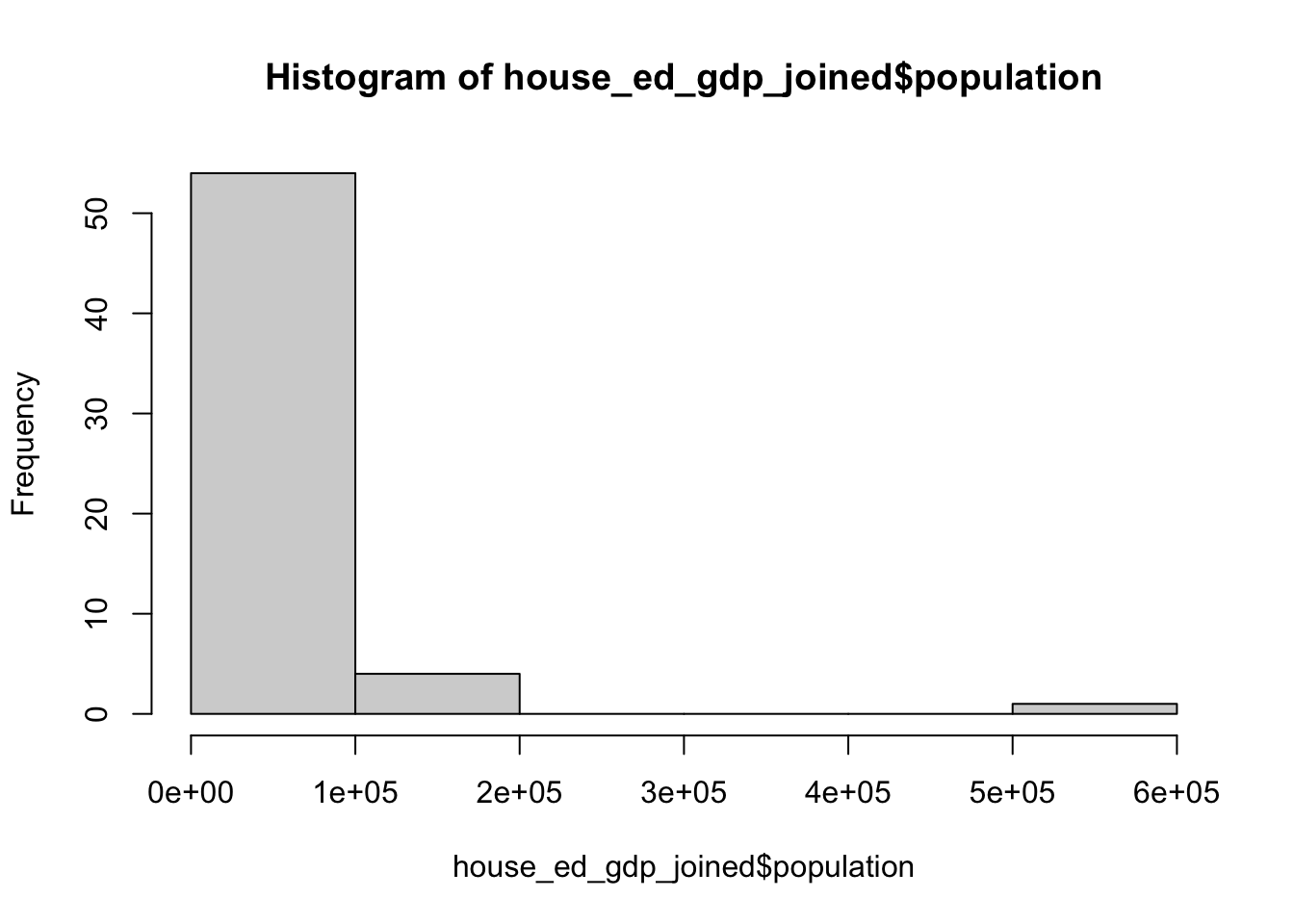
counts <- table(house_ed_gdp_joined$culture_site) # This counts our variable by category
barplot(counts, main = "title", xlab = "x label", ylab = "y label") # a bar plot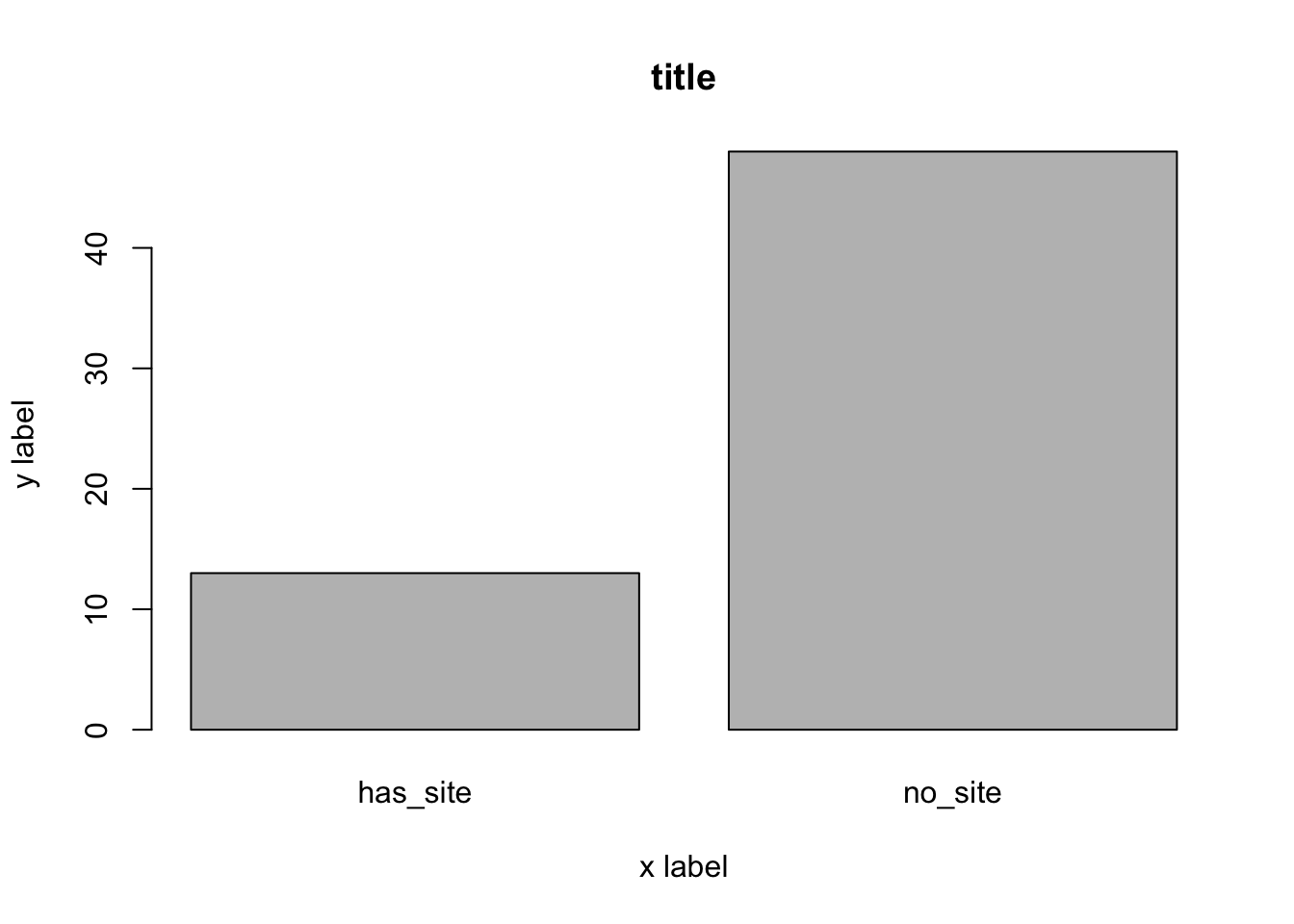
You can see that this does the job, but it is not very inspiring. Even if you were to take the time to give it better axis labels, it would still look basically the way it does right now.
So, let’s use tidyverse to get a bar plot:
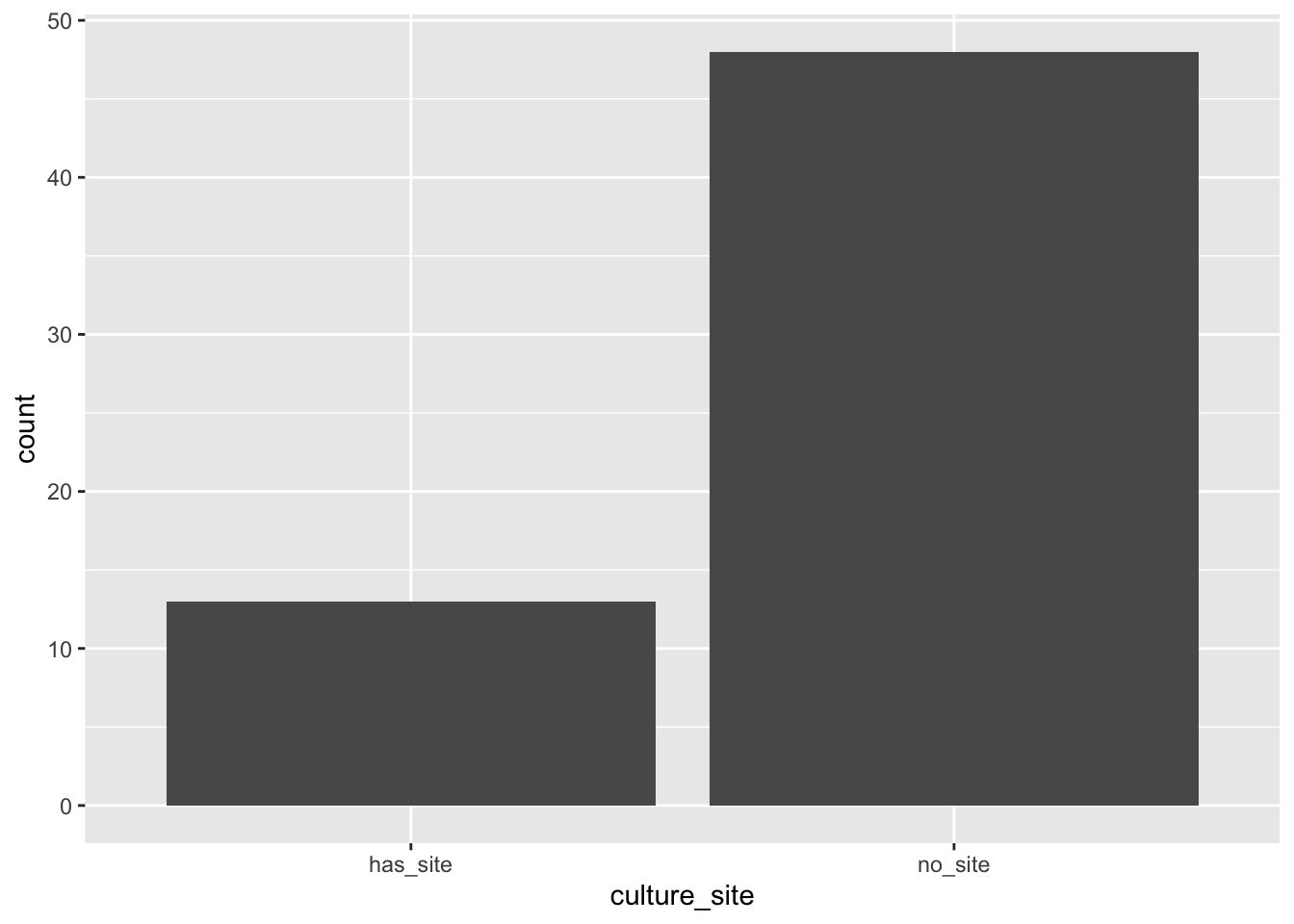
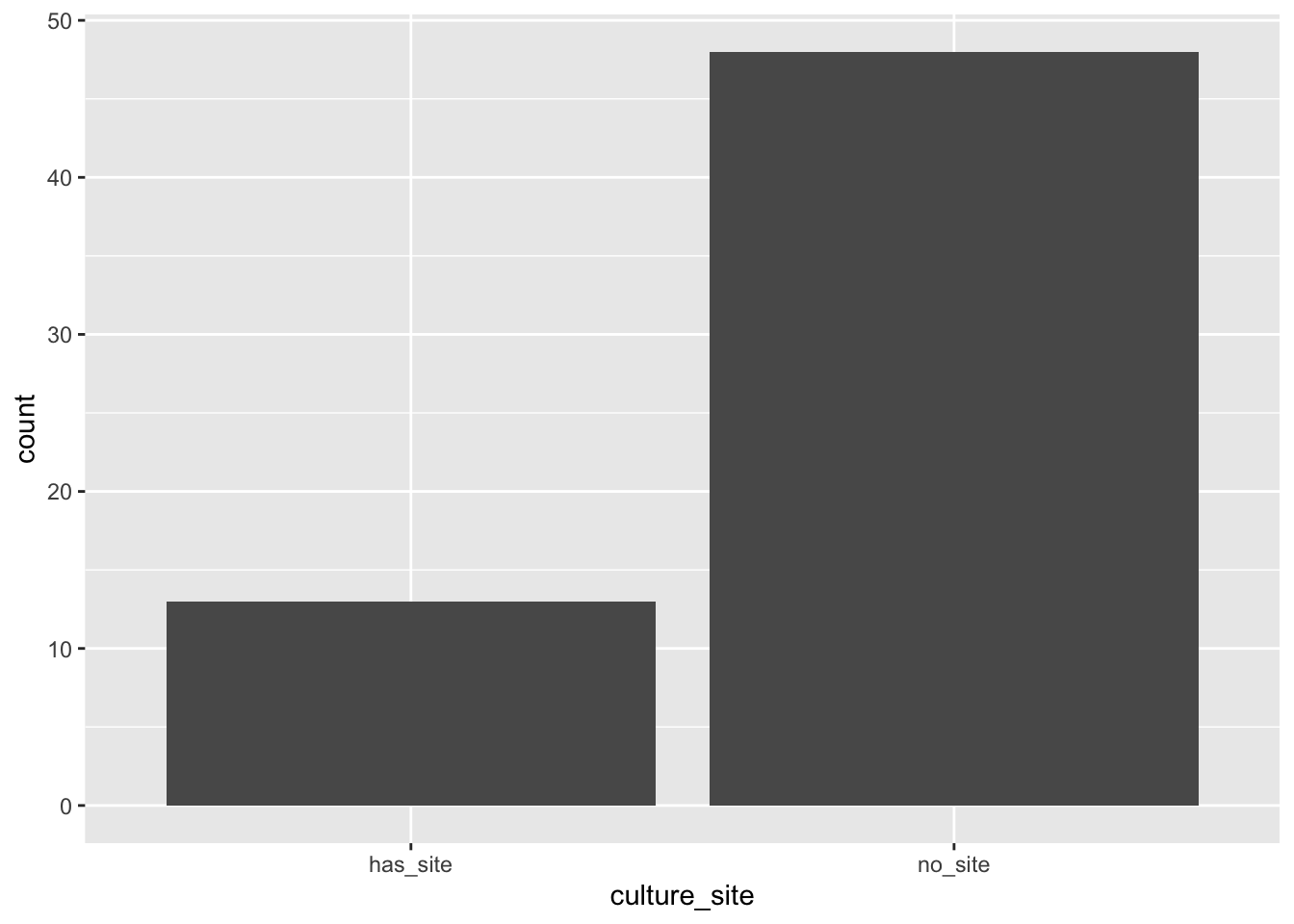
Notice that either geom_bar or stat_count returns the same bar plot.
Tidyverse has a series of “geoms” or visual tools. It also has “stats” or numerical tools. The geom geom_bar is paired by default to the stat stat_count. Every geom is paired to a default stat and vice versa. As a convenience, you can use the name of the default stat to use its geom. This may seem confusing, but it is actually convenient. If you wish to override the defaults, use optional arguments to these functions.
https://rstudio.com/wp-content/uploads/2015/03/ggplot2-cheatsheet.pdf has a nice reference grid.
If you wish to see the various geoms or stats in RStudio, type geom and wait there at the end of the m (or stat and wait at the end of the t). A menu should appear to guide you through the information you seek. To explore a specific geom or stat, type ?gemo_bar (etc).
3.5 Variations on bar plots
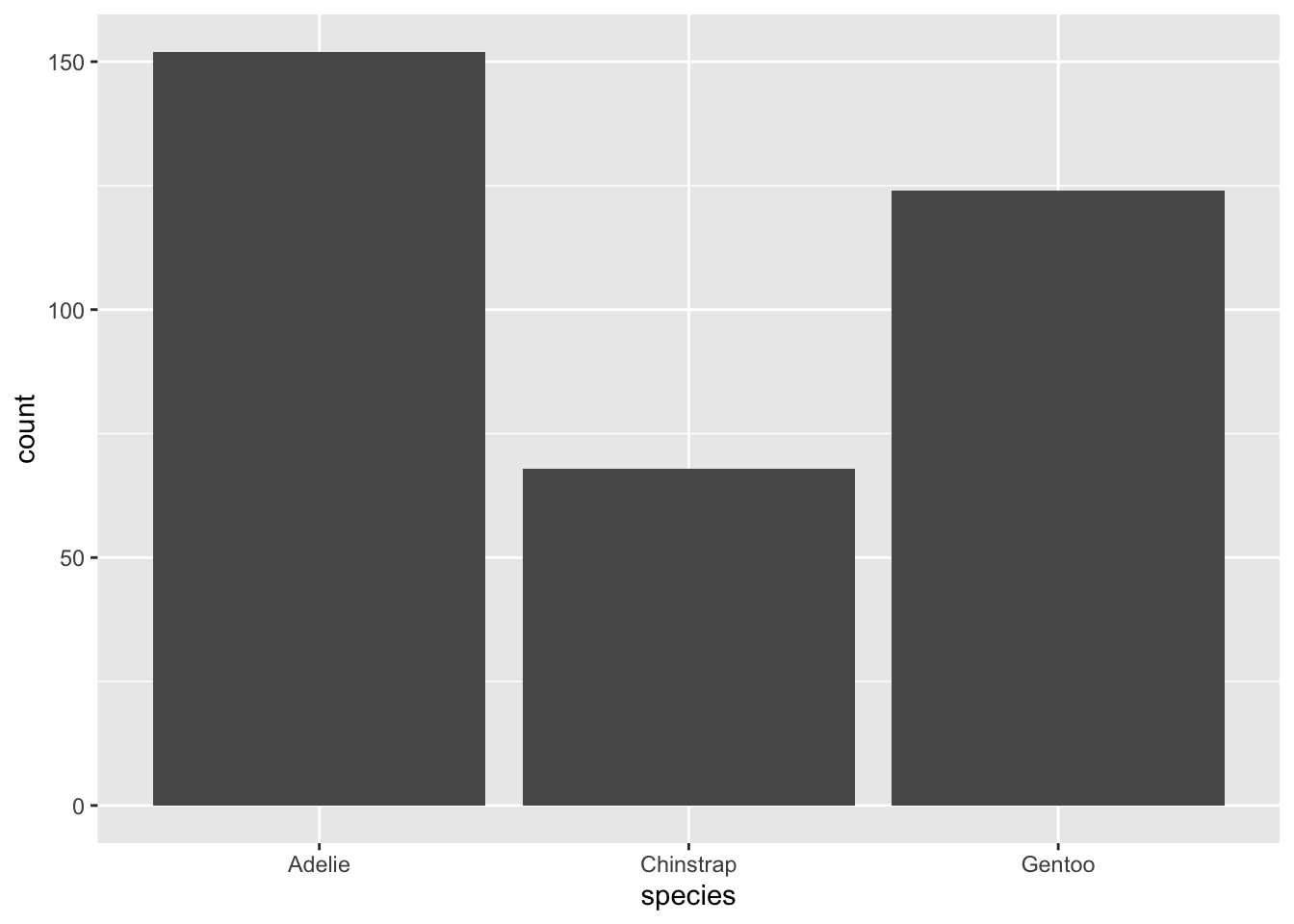
Let’s switch over to the penguins! The dataset contains observations of various penguins.



How many observations?
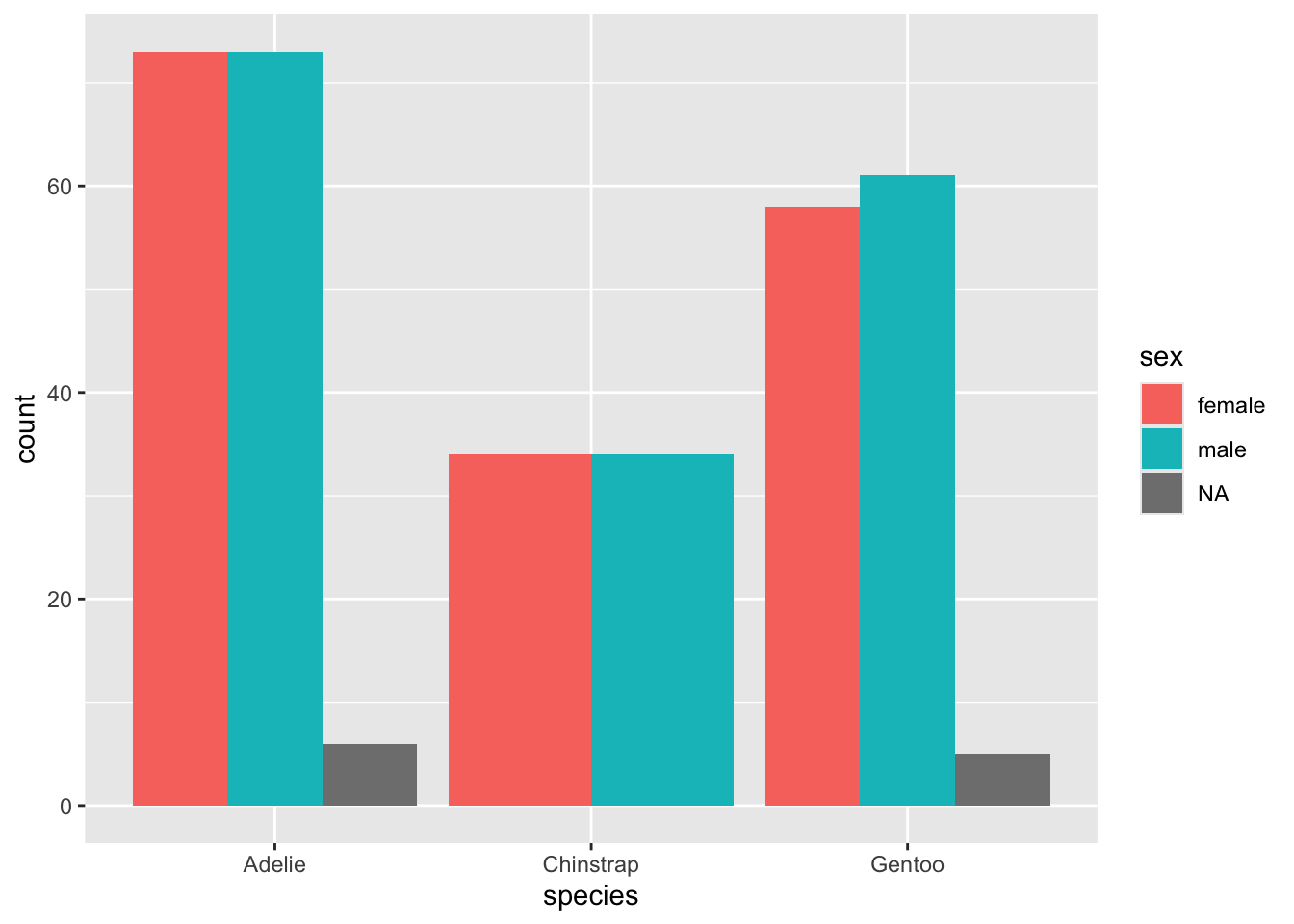
We have not run into column names with spaces in them. You can use back-quotes (on US keyboards, to the left of the number 1) to enclose the full column name. For instance, here is a grouped histogram…
But not every observation is a different bird. There’s a column Individual ID that identifies the different birds. Here’s how many birds there are:
And finally, an obligatory summary!
species island bill_length_mm bill_depth_mm
Adelie :152 Biscoe :168 Min. :32.10 Min. :13.10
Chinstrap: 68 Dream :124 1st Qu.:39.23 1st Qu.:15.60
Gentoo :124 Torgersen: 52 Median :44.45 Median :17.30
Mean :43.92 Mean :17.15
3rd Qu.:48.50 3rd Qu.:18.70
Max. :59.60 Max. :21.50
NA's :2 NA's :2
flipper_length_mm body_mass_g sex year
Min. :172.0 Min. :2700 female:165 Min. :2007
1st Qu.:190.0 1st Qu.:3550 male :168 1st Qu.:2007
Median :197.0 Median :4050 NA's : 11 Median :2008
Mean :200.9 Mean :4202 Mean :2008
3rd Qu.:213.0 3rd Qu.:4750 3rd Qu.:2009
Max. :231.0 Max. :6300 Max. :2009
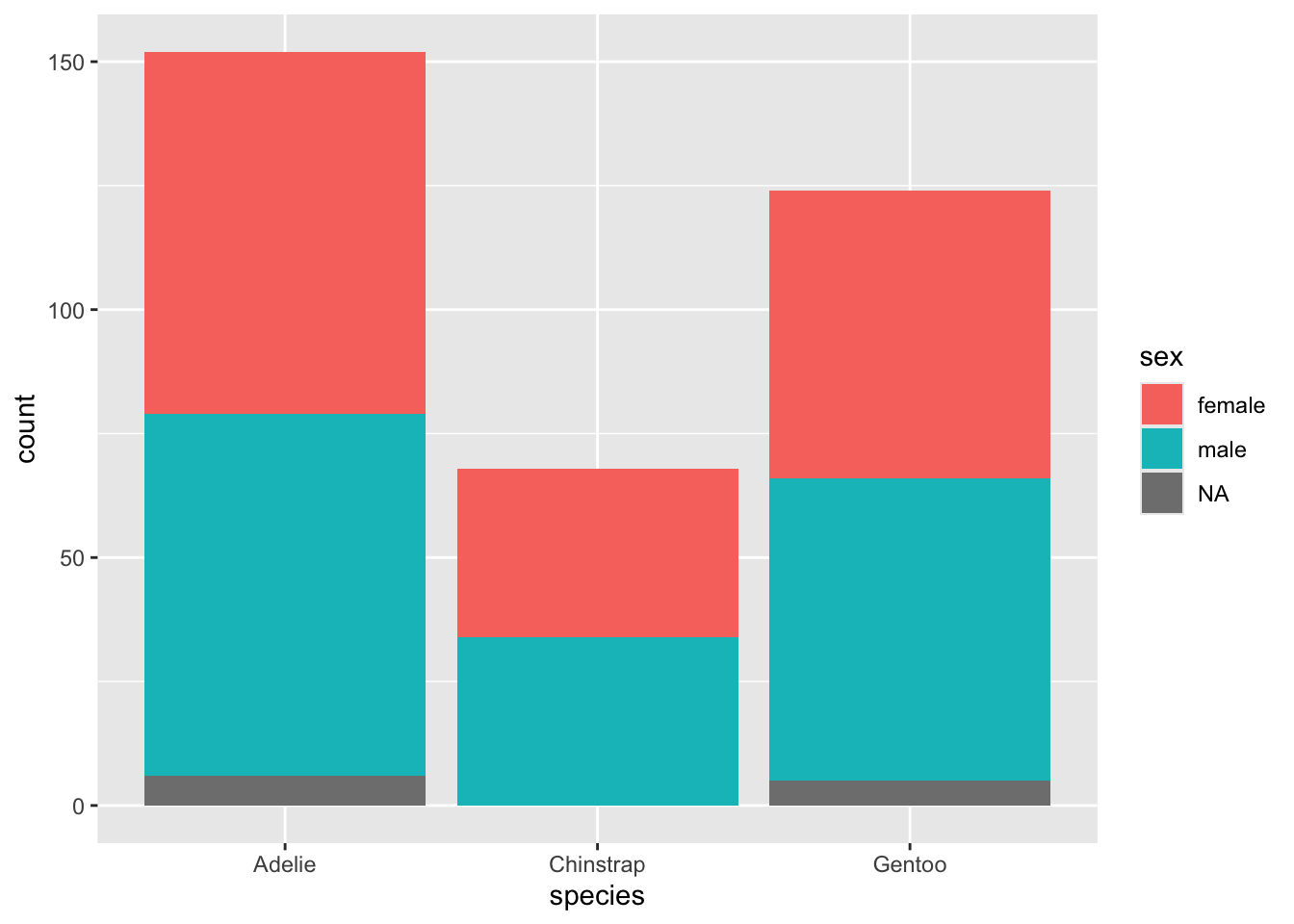
NA's :2 NA's :2 You can also use the coloring to explore another categorical variable within your categories.
Perhaps you wish to line up all the categories by their subcategories, each in individual bars.
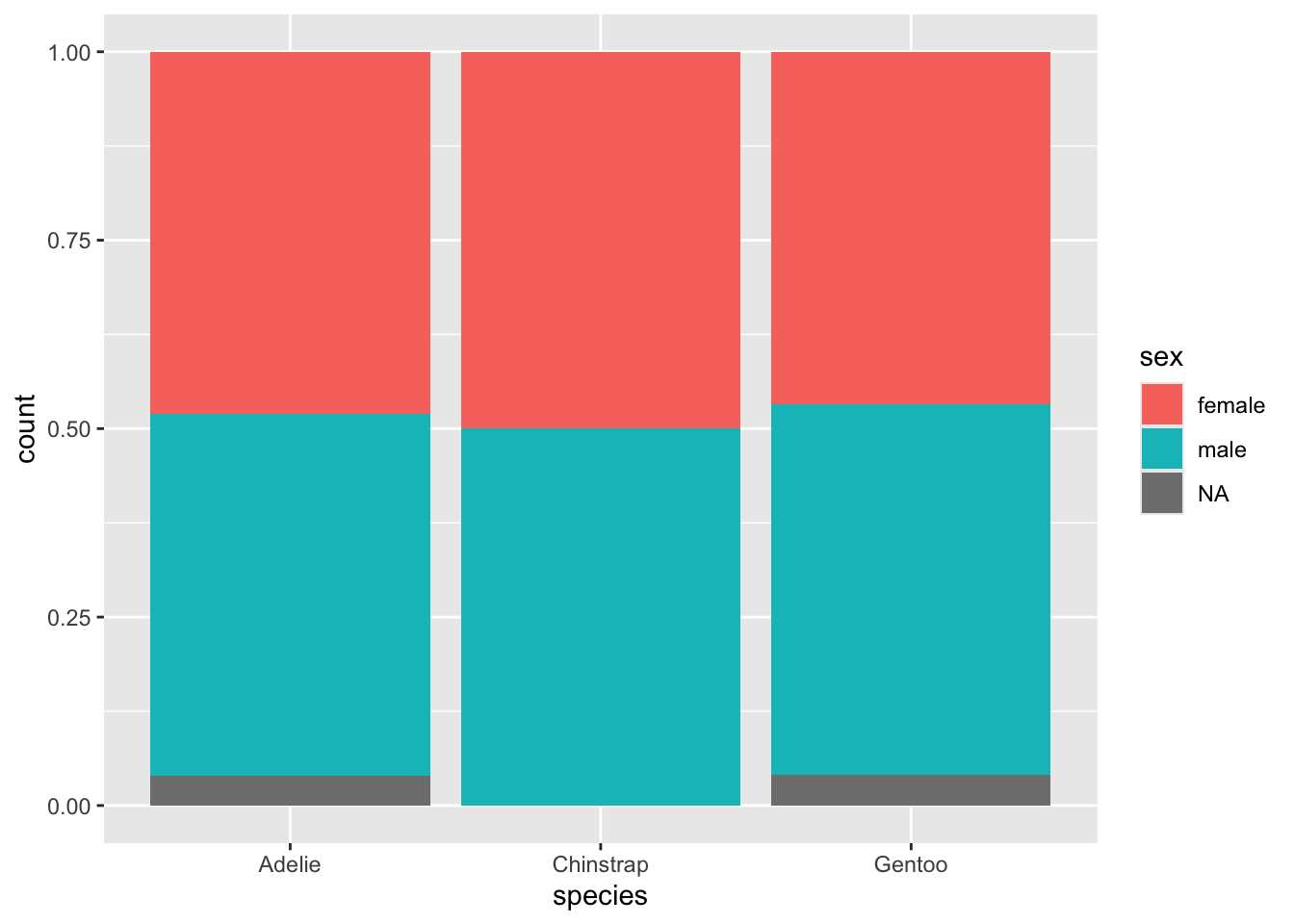
Or, perhaps you prefer to weight each of your primary categories to 100% so you can visually see if the proportions of the second category are roughly the same or not.
3.6 Pie charts (round relative frequency histograms)
The usual pie chart can be drawn as follows and makes use of the same table command we already saw above for bar plots.
mytable <- table(penguins$species)
myLabels <- names(mytable) # You could change the labels here.
pie(mytable, myLabels, main = "species")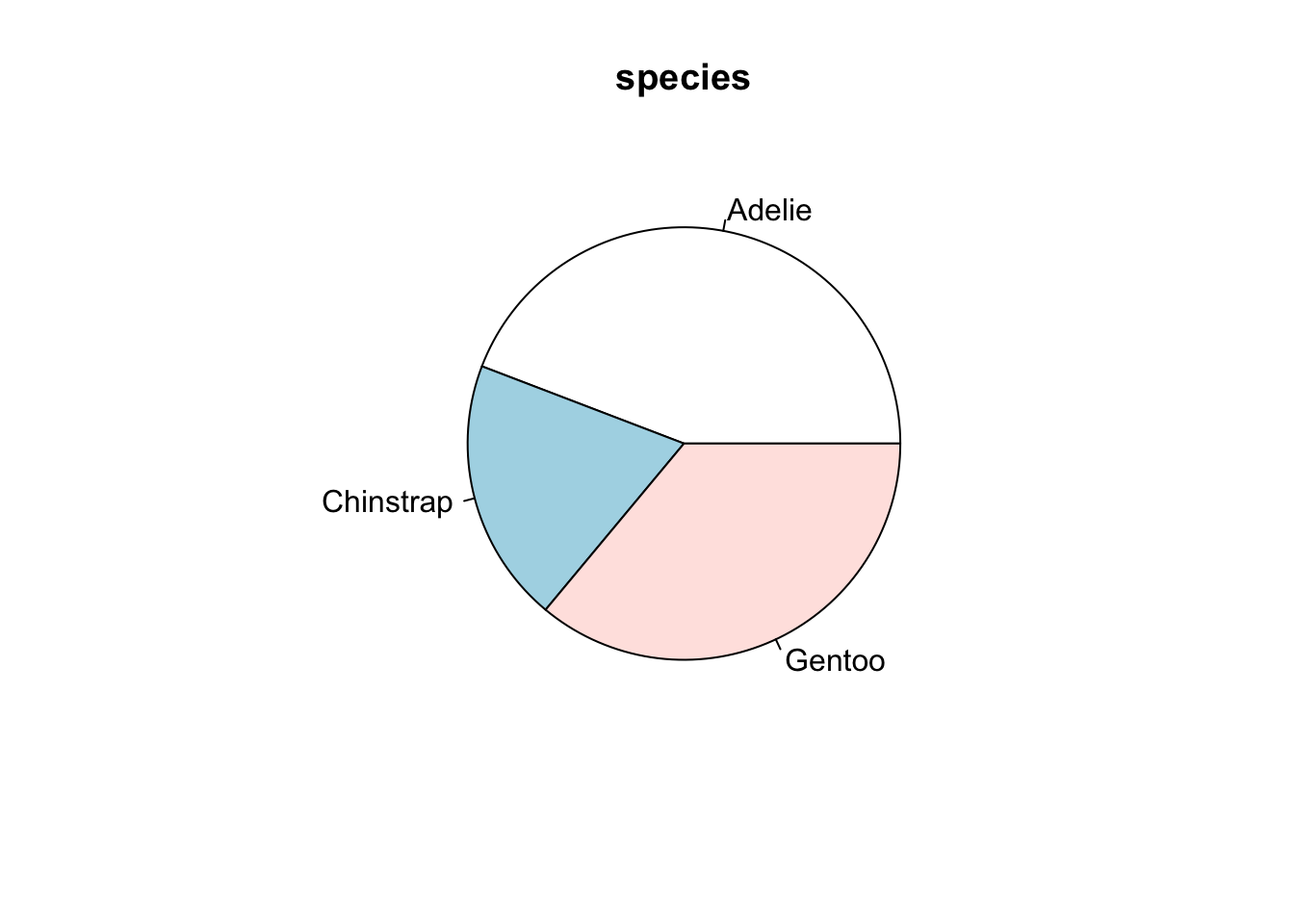
A tidyverse pie chart looks quite different and is more customizable. But, it takes many more steps. Here, we follow the instructions from: https://www.tutorialspoint.com/ggplot2/ggplot2_pie_charts.htm
df <- as.data.frame(table(penguins$species))
colnames(df) <- c("jobs", "freq")
pie <- ggplot(df, aes(x = "", y = freq, fill = factor(jobs))) +
geom_bar(width = 1, stat = "identity") +
theme(axis.line = element_blank(), plot.title = element_text(hjust = 0.5)) +
labs(fill = "jobs", x = NULL, y = NULL,
title = "Pie Chart", caption = "Caption")
pie # Look at pie now! After all that effort, it's not even a pie yet!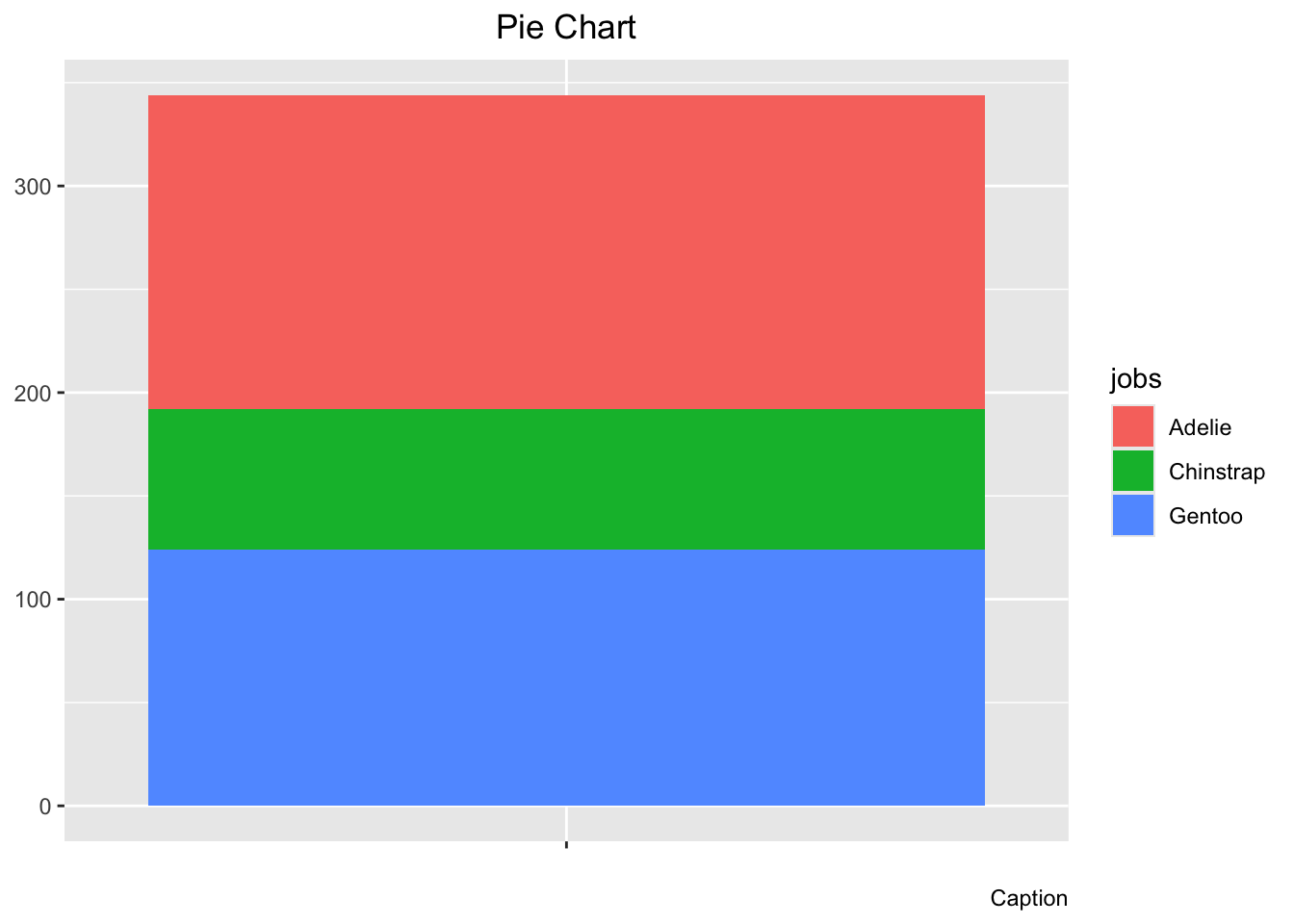
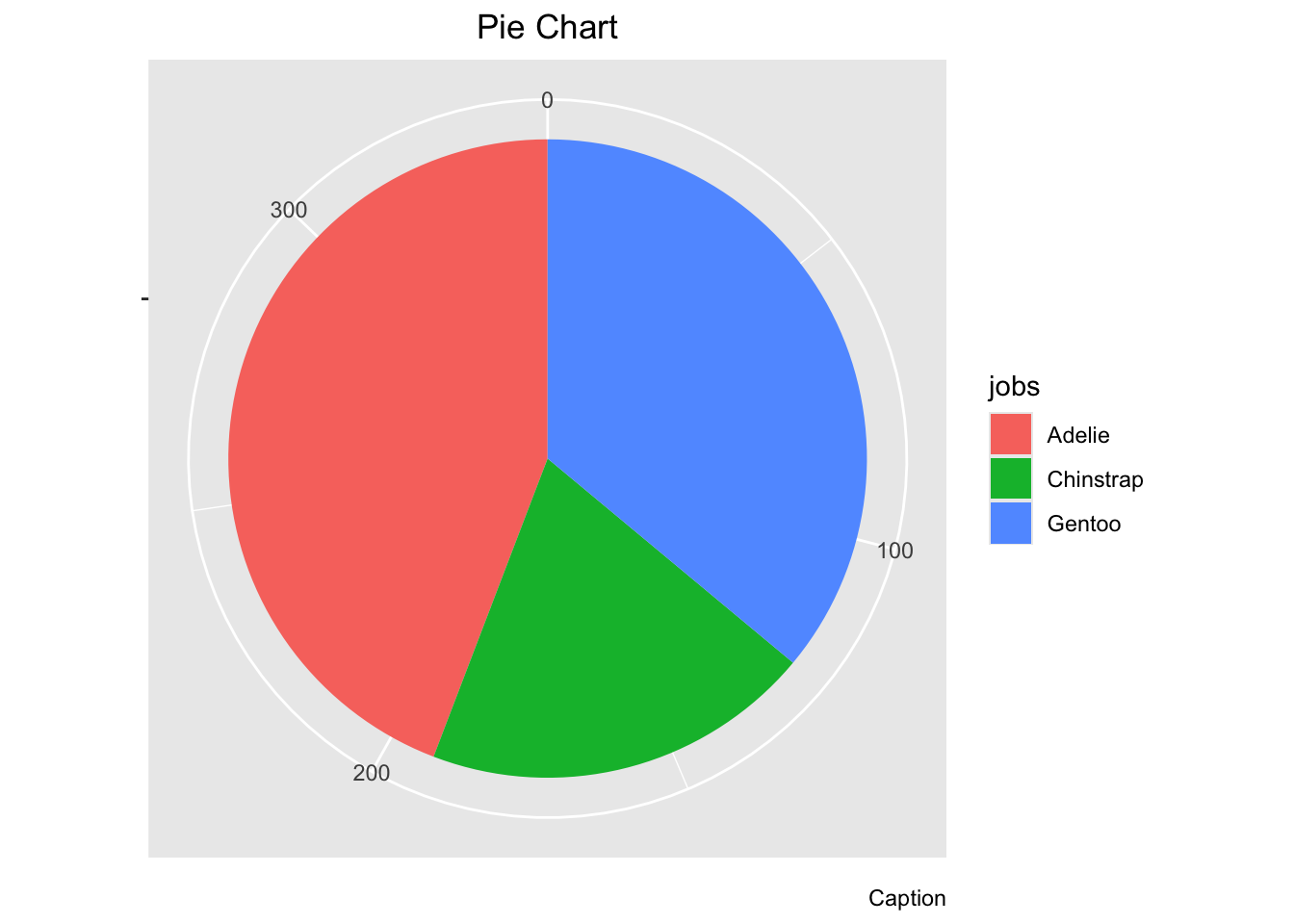
3.7 Comparing distributions
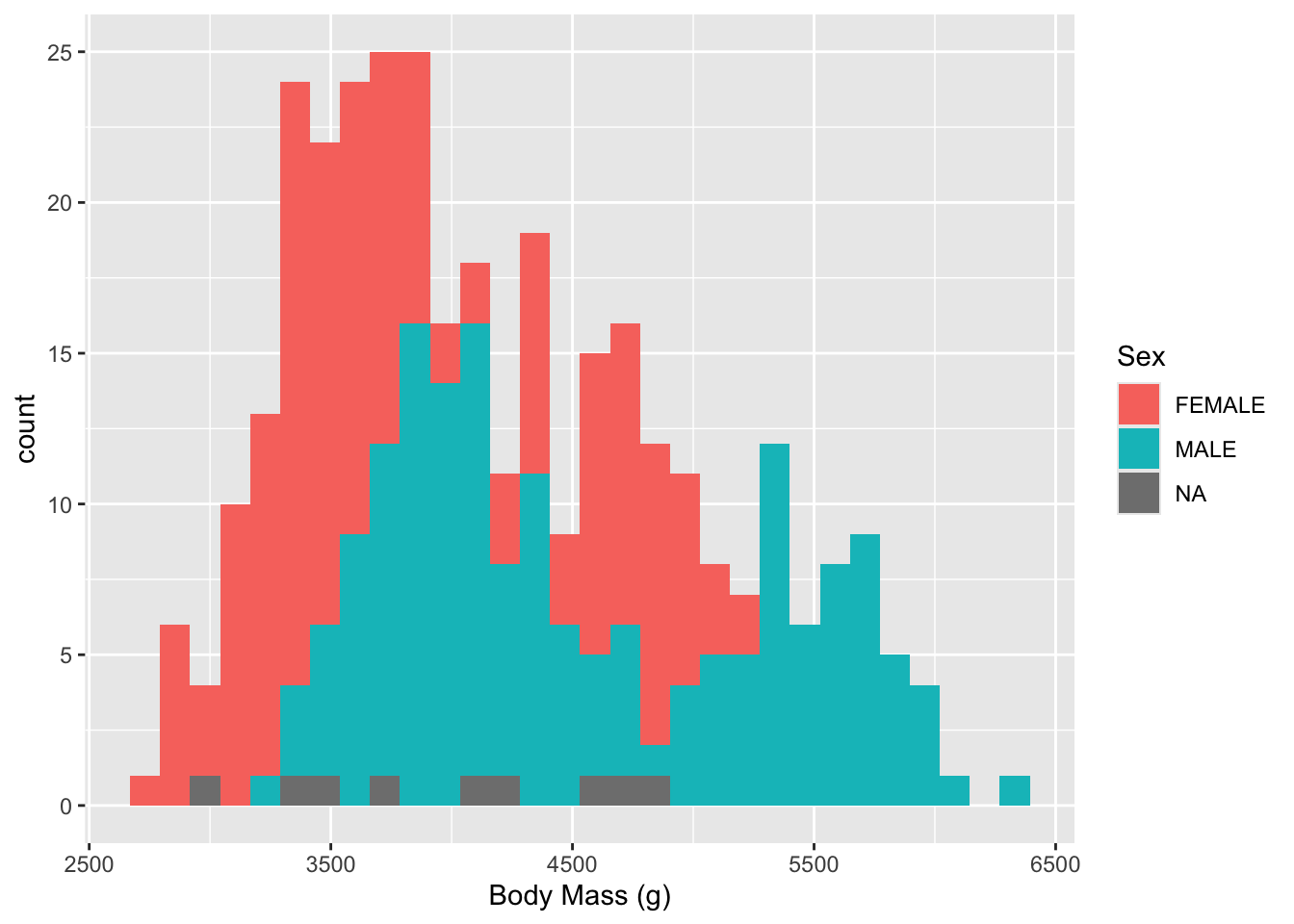
Let’s get a histogram of the mass observations (noting that there may be multiple observations from each bird as it grows)
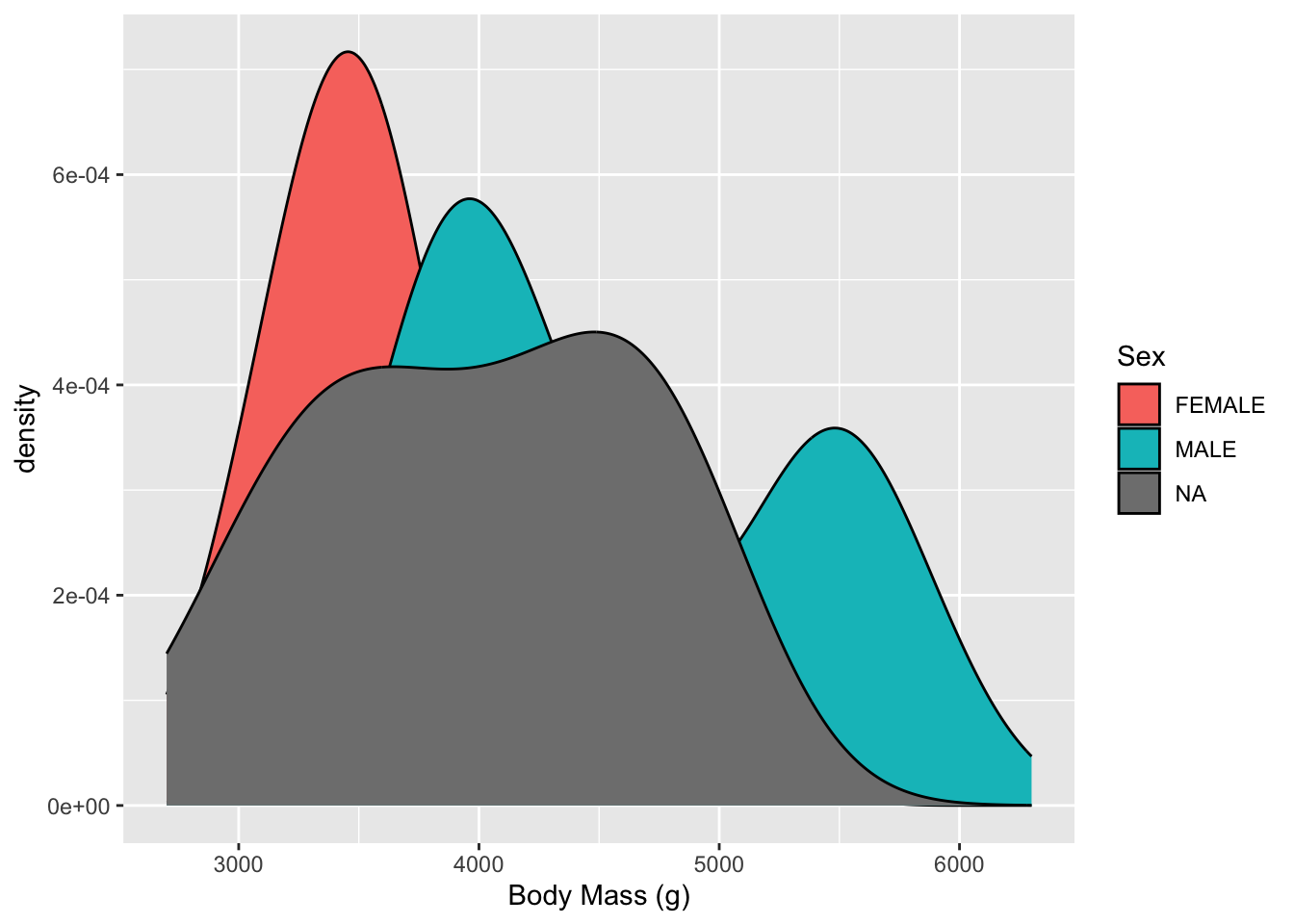
Another view is a Density plot. This is a smoothed version of the histogram.
- All the NAs get in the way so let’s get rid of them.
penguins_raw |>
filter(!is.na(Sex), !is.na(`Body Mass (g)`)) |>
ggplot(aes(fill = Sex, `Body Mass (g)`)) +
geom_density()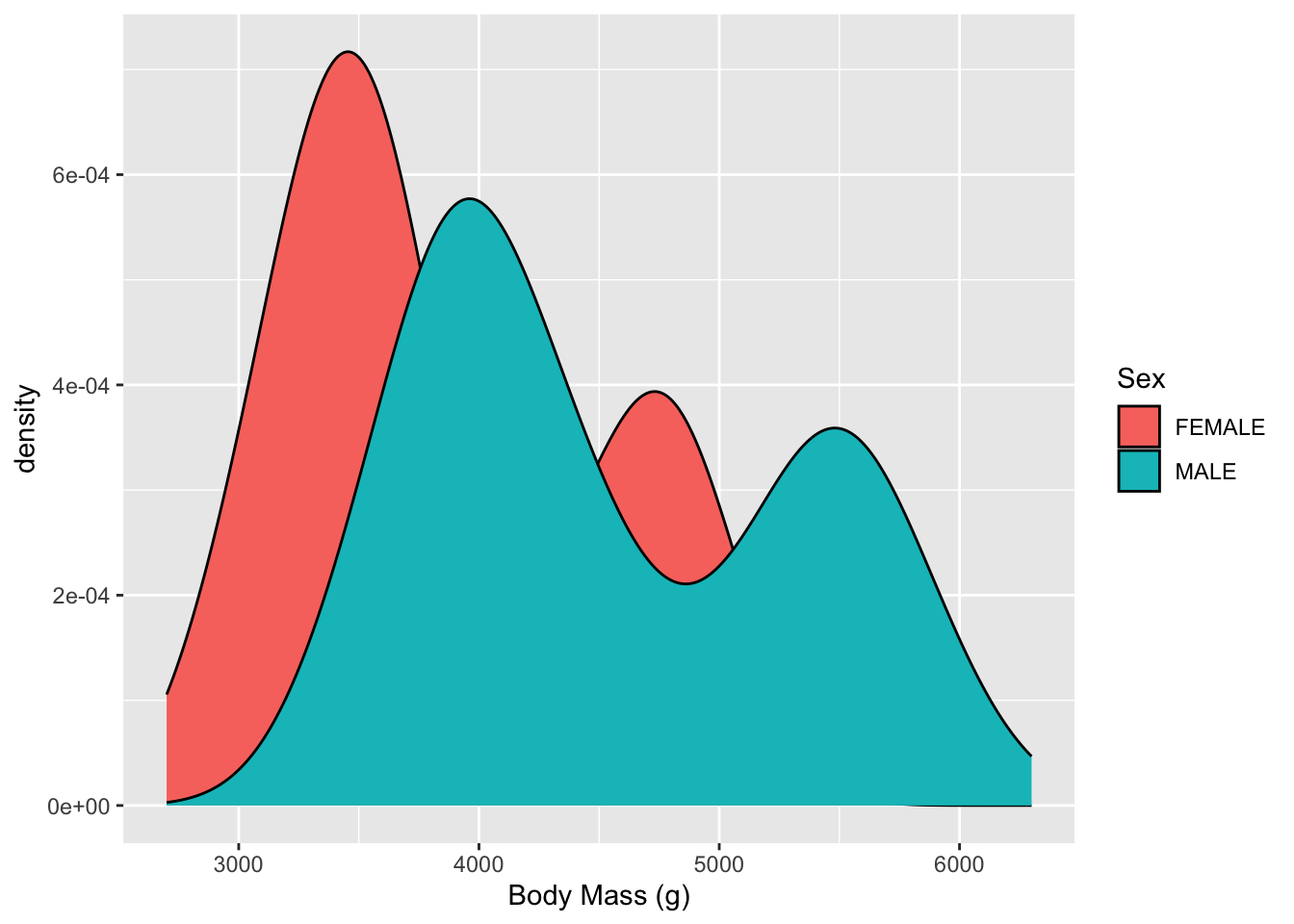
- Now we can;t see everything so lets increase the transparency (decrease the opaqueness)
penguins_raw |>
filter(!is.na(Sex), !is.na(`Body Mass (g)`)) |>
ggplot(aes(fill = Sex, `Body Mass (g)`)) +
geom_density(alpha = .2)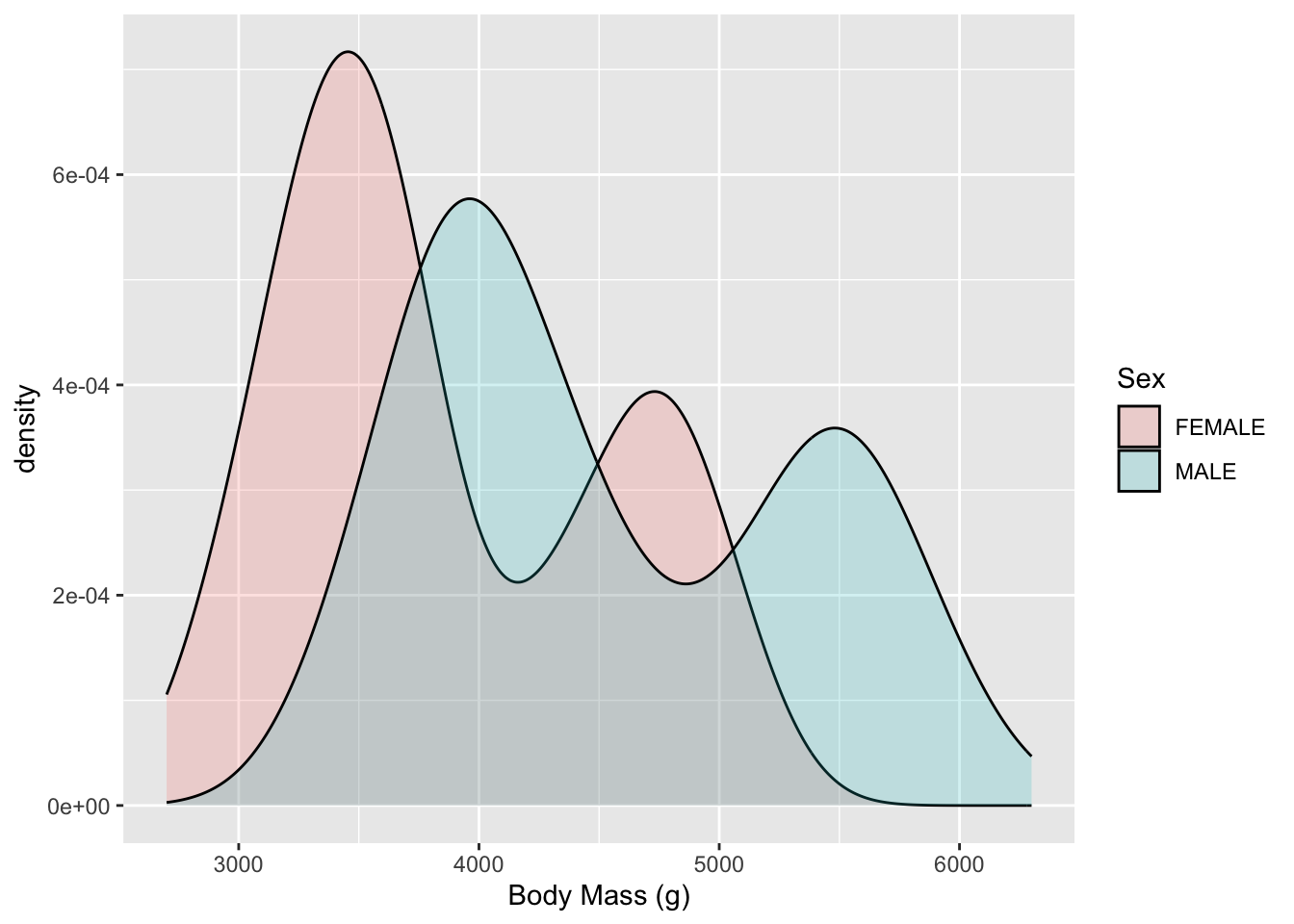
- Now we can see both Males and Females have two humped plots or what we call Bimodal (the mode is the most common value).
- This suggests that the distributions for each sex are actually a mixture of two different kinds of makes and females.
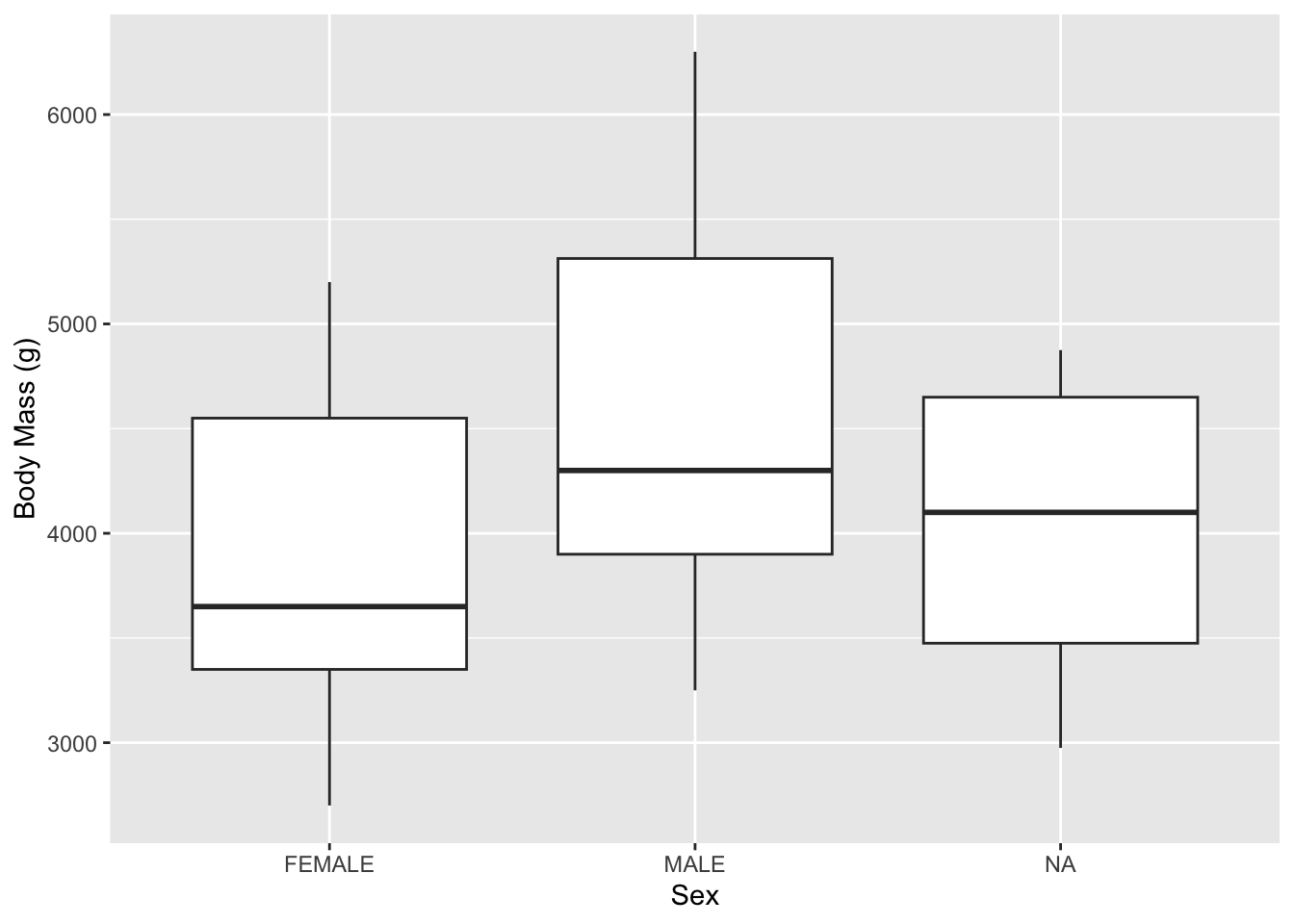
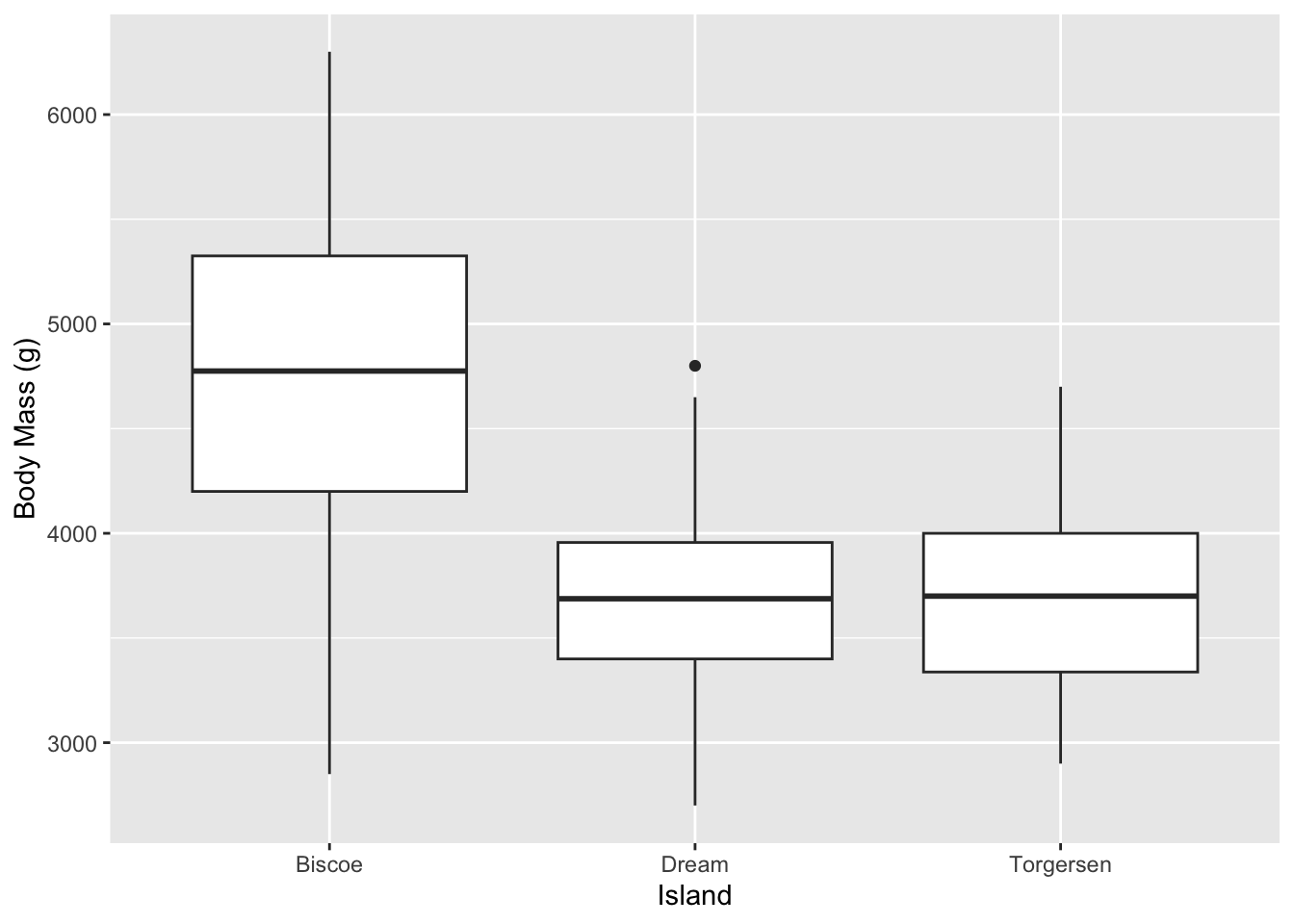
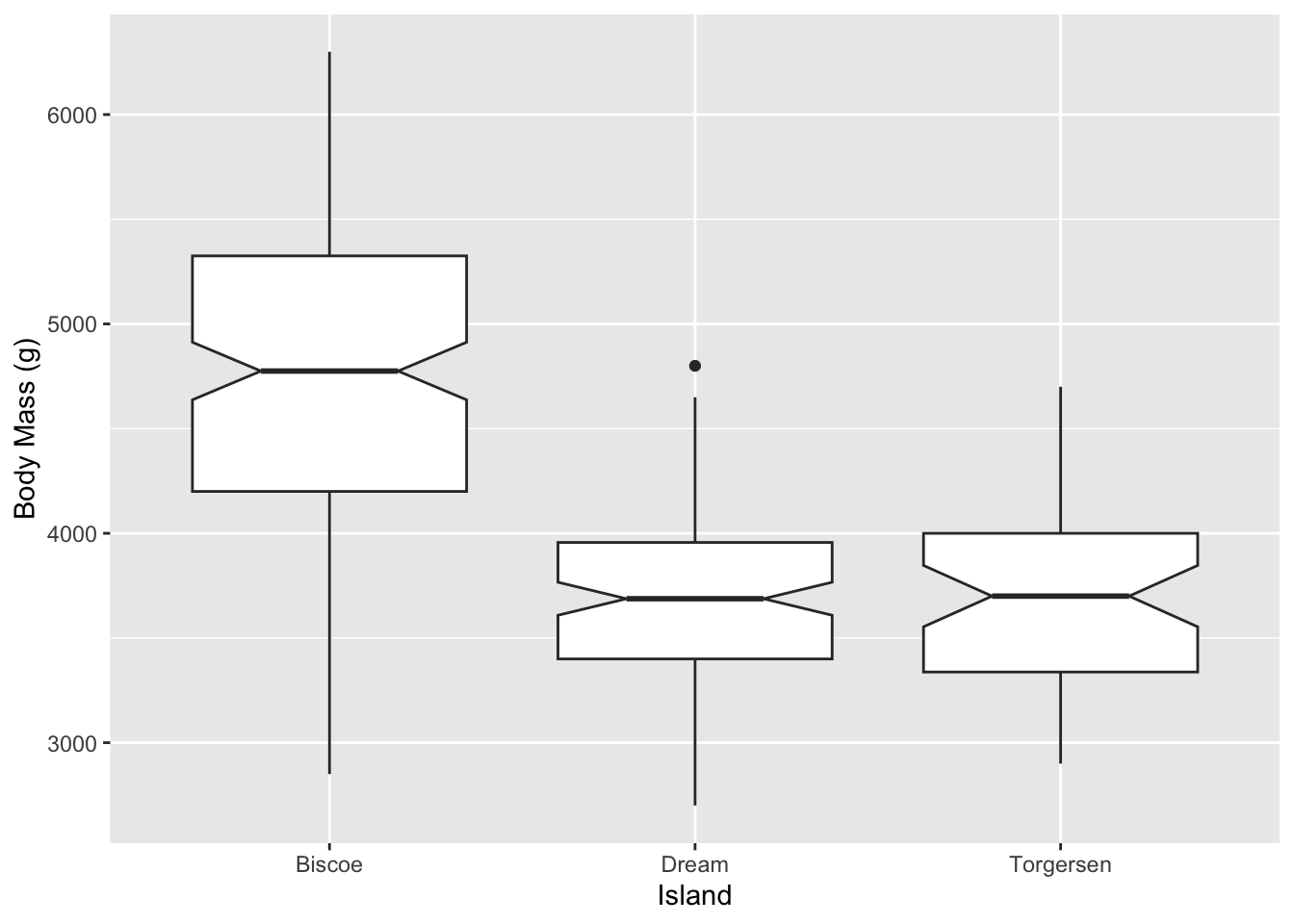
But maybe that is too busy of a plot. We can clean that up by using other geom_ choices. For instance, here is a classic box plot:
Notice that the aes() which selects which variables are involved is a little different between the above two commands. You’ll often need to consult the documentation to see what the choices are.
Now if you set the notch = TRUE argument in geom_boxplot() you will some notches appear.
- As the help says for the
geom_boxplot()notchargument “Notches are used to compare groups; if the notches of two boxes do not overlap, this suggests that the medians are significantly different.
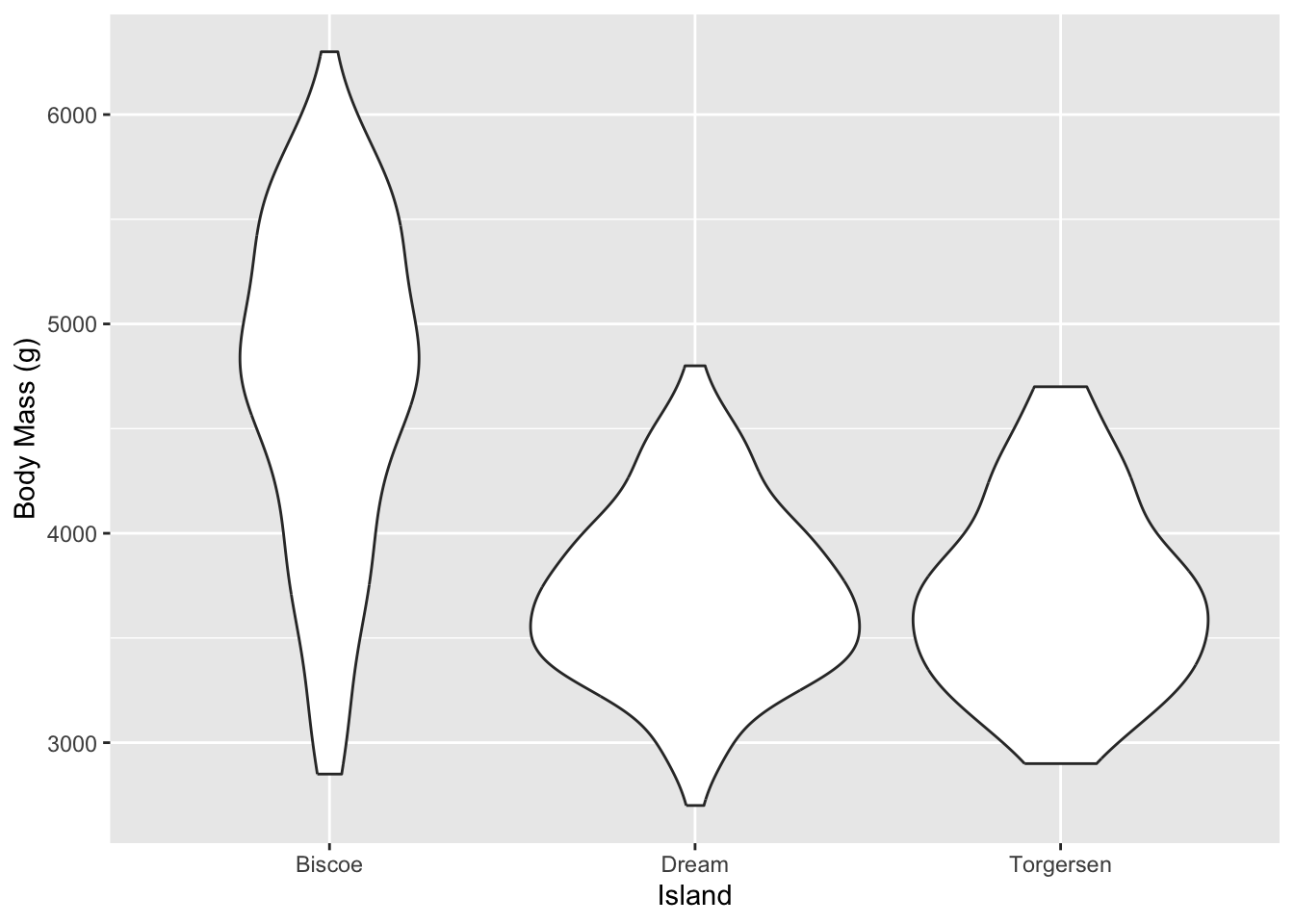
Another variant is the Violin plot. This combined the box plot with the histogram density plot.
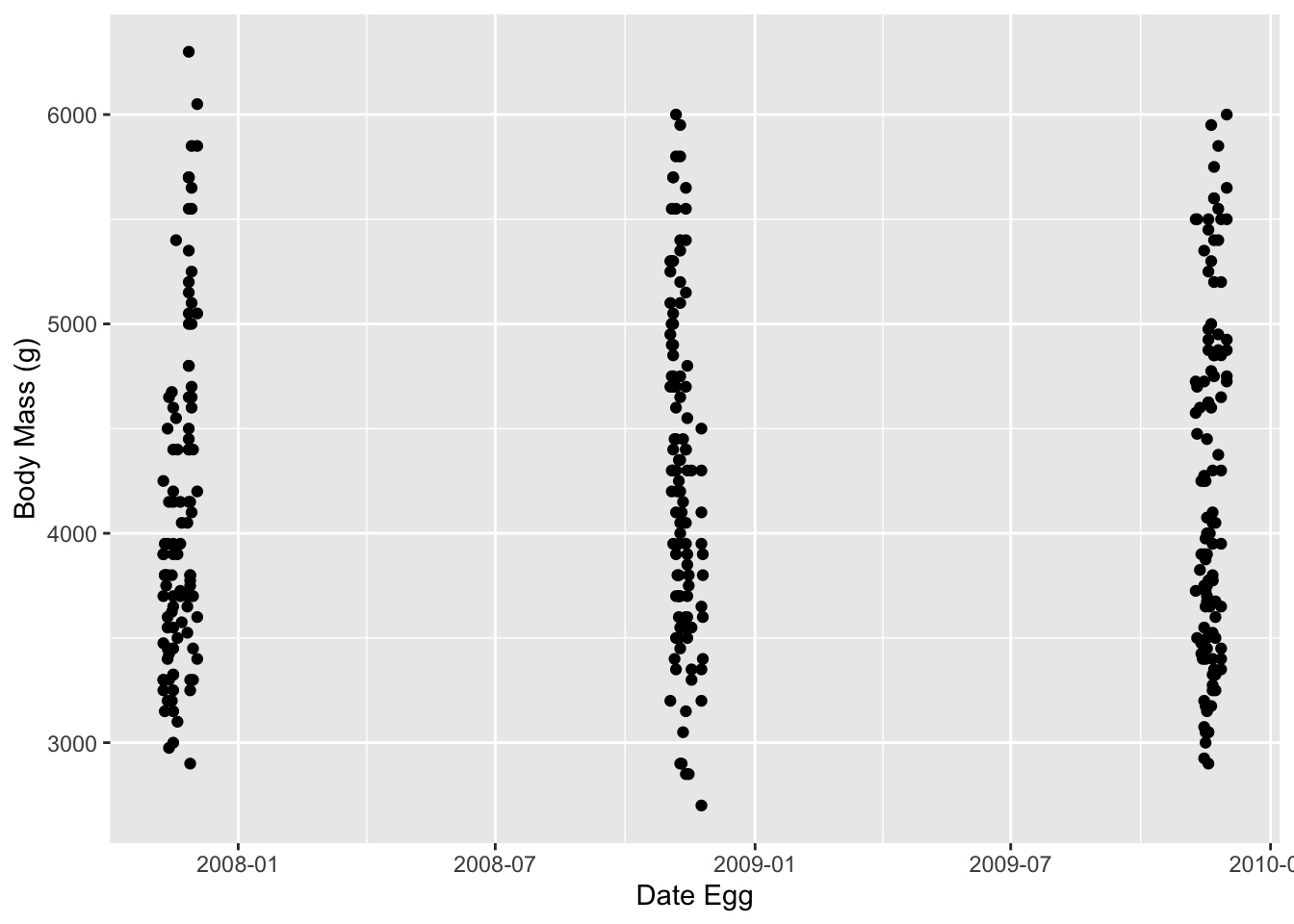
Although we’ll work with scatterplots more next lesson, here’s a taste of what you can do:
It looks like the dates came in bunches, probably when the researchers were out looking at the penguin nests.
3.8 Exercise
- Select a dataset
You can use the same dataset for Lessons 3, 4, and 5. In your data set, you should have at least two variables (columns) which can be interpreted as categorical and at least two other variables which are numerical in nature.
- Classify all your variables as numerical or categorical.
Binary variables are a special kind of categorical, as are factors (ordinal variables). We are not making a distinction between continuous or discrete numerical variables, except for this: If you have too few numbers, you should suspect that the variable might be better classified as a categorical variable. For example, age is often a variable that could go either way, especially if your data concerns children who are close in age.
- Demonstrate basic descriptive statistics
For one or more numerical variables, find the mean, mean, variance, and standard deviation.
- Make a histogram
Use one of the numerical variables! You might find it interesting to change the fill or group based upon a categorical variable.
- Make a bar plot
Bar plots are made from grouping categorical variables. Explore a few of these.
- Create a pie chart
Demonstrate the use of a Pie chart.
Do one of the following, using the same two categorical variables you used for your Chi-Square test.
- Using subsets, split your data into categories, and create a pie chart for each group, based on another categorical variable in your data.
- Use
geom_barwithposition="fill"to create side-by-side bar plots that demonstrate the ratios occupied by a second categorical variable.
- Create another plot!
Choose from geom_boxplot(), geom_jitter(), or many others! Read the documentation to find some options!
- Descriptive work
Since you’re working with Quarto, you can write some text between your results. Explain your findings! Show the person next to you what you found most interesting.