2 Cleaning and Reshaping Data
data wrangling, dplyr, tidyr, stringr, regex, joins
2.1 Introduction
This module focuses on step 4 in the life cycle- Shape Data. It introduces some core data science skills in cleaning and shaping data - it’s what data scientists do.
Learning Outcomes
- Explain the need to clean data.
- Clean and shape rectangular data: filter rows, select columns, rename columns, …
- Explain basic methods for reshaping and combining data: pivots and joins.
2.1.1 References
These are all core {tidyverse} packages:
- Manipulating Data Frames: {dplyr} package (Wickham et al. 2023)
- Manipulating String or Character Data: {stringr} package (Wickham 2023c)
- Manipulating Dates and Time Data:{lubridate} package (Grolemund and Wickham 2011)
- Manipulating Factor or Categorical Data:{forcats} package (Wickham 2023a)
- Reshaping Data Frames: {tidyr} package (Wickham, Vaughan, and Girlich 2023)
Other References
- Cleaning Data Frames: {janitor} package (Firke 2024)
2.2 Cleaning Data
Once we have a question, framed the analysis, and found some data, it is time for step 4 in the life cycle - cleaning and shaping the data so it can be used for analysis.
2.2.1 Real Data is Messy
Data from formal research experiments with randomized trials is usually well-designed to support the analytical approach. (Thank your statistician friends!)
- There can still be measurement errors, input errors (Female, Femmale, Fem, 1, Woman), sampling errors, multiple ways of indicating
NA(na, -999, null) or duplication.
However, most data is “observational”, created for purposes other than your analysis.
- Even with the growth of the Internet of Things (IOT) devices generating data, humans still create more data than devices, and most of that data is text.
Observational real-world data is usually messy. In addition to the errors in “good” experimental data, …
- It is often incomplete, so not every variable is present that you might want.
- Humans are “notorious” for their creativity when using text - it is a feature and a bug.
- Even when entering numeric data, we make mistakes such as switching numbers, adding spaces, putting the decimal point in the wrong place, ….
- Most data is designed for humans to read and not for computers to analyze.
- Real-world data is often missing values, has different formats (e.g., times and dates).
- When data is scraped from web pages or PDFs there are often random errors in it.
- Tabular data in customized tables with merged rows or columns can be hard to parse correctly.
Given the vast amount of messy data, there are multiple names to describe what happens in cleaning and shaping data. These include
- Data “wrangling”: The overall process of cleaning, transforming, and preparing raw data for analysis.
- Data “munging”: Informal synonym for data wrangling; often used to describe converting messy or unstructured data into a usable format.
- Data pre-processing: Steps taken before actual analysis, such as handling missing values, encoding categories, and scaling.
- Feature engineering: Creating new variables (features) or modifying existing ones to improve the performance of models.
- Data normalization: Transforming data to a common scale without distorting differences in the ranges of values (e.g., min-max scaling or z-score standardization).
Regardless of the name, the steps of getting the data and then preparing it for analysis often take much longer than and require as much creativity as building a model or running statistical tests.
Surveys support the idea that many data scientist spend between 40% to 80% of the time on a project working to get data, clean it, and shape it.
- This is often an iterative process as you try some data and then realize you need more.
2.3 Rectangular and Tidy Data
The goal for cleaning and shaping data is use a reproducible process to get the data in its most accurate and consistent representation in a format that is easy to use in analyzing the question of interest.
- We want reproducible process so we can more easily update the data if new data arrives, say a new month or year of data is released.
- We want the data to be accurate and consistent so we minimize noise in our analysis.
- We want the data in an easy to use format so we can apply standard methods and tool.
Starting with the format, we want our data in rectangular format known as a data frame.
- R, Python and Excel all work best with rectangular data: a table where the rows are the multiple observations (records) for an entity and the columns are the attributes of the entity which we refer to as variables.
In R, a data frame has class data.frame and the tidyverse works with a special kind of data.frame known as a tibble.
Both data.frames and tibbles though can have messy data in them, a prime example is where several variables have been “mushed” together for human use or have been spread out across the columns, e.g. a different month in each column. This hides the fact that month is actually a variable with the value of the month.
Thus the tidyverse also uses the term “tidy” to describe a data frame or tibble where “Each variable forms a column, each observation forms a row, and each type of observational unit forms a table”.
In R a data.frame is actually a special case of a list which is a type of vector.
- An R vector is a one-dimensional object that has one or more elements in it. Each element has a class. The length of the vector X,
length(x)is the number of elements in it. - There are two kinds of vectors: Atomic and List.
- For a vector to be Atomic, all elements must have the same class, e.g., numeric, logical, factor, character, or even complex or raw.
- If the vector is Not atomic, it is a list. That means it can have elements with different classes. A list can also have elements that are atomic vectors or lists themselves so it can have many levels and each element may have a different length.
- However, the length of a vector is always the number of elements at the top level.
- A data.frame is is a special kind of list where every element does not have to have the same class, but all elements must be named atomic vectors with the same length.
- When we look at a data.frame or tibble in R, think of each column as a named atomic vector that has length
n, the number of rows.
- When we look at a data.frame or tibble in R, think of each column as a named atomic vector that has length
- R vectors can be combined into other special classes such as matrices which can have multiple dimensions of atomic vectors or time-series class which are vector with special attributes for start time, end time, and frequency.
The length of a data frame is the number of columns, use length() or ncol() and to get the number of rows use nrow().
For this course we will generally work with atomic vectors (usually just called vectors) and data frames although a list may pop up from time to time. You will see these terms in the help documents and perhaps the error messages so now you have some context for them.
2.4 Using Tidyverse Packages to Clean and Shape.
The Cheat Sheets provide a quick summary of the important packages
- Data transformation with dplyr
- String manipulation with stringr
- Dates and times with lubridate
- Factors with forcats
- Tidying data with tidyr
Each of these packages also have extensive documentation to include vignettes at their websites references in Section 2.1.1.
Instead of going through the functions per se, let’s get some data and see what we need to do to get the data clean and tidy.
2.4.1 Load Data Sets
Let’s start by loading the {tidyverse} set of packages.
The data folder has several data sets. These include
Data from Open Data Albania (https://opendata.gov.al/) required translation (used ChatGPT and Claude)
- museums_raw.csv: Number of foreign and domestic visitors in parks, museums, and cultural centers for the period November 2018 - August 2019 data from the Ministry of Tourism and Environment
- gdp_per_capita_2023.csv: Gross Domestic Product per Capita 2023 data from the Ministry of Finance
- schools_raw.csv: Secondary Schools data from the Ministry of Education, Sports and Youth
Data from INSTAT (https://databaza.instat.gov.al:8083/pxweb/en/DST/)
This data did not require translation but it did require exporting as XLSX to preserve the diacritical marks which were corrupted when exported to CSV.
education.xlsx
households.xlsx
sex_age.xlsx
Let’s load the open data files.
Before loading the XLSX files, we need to look at them to figure out their structure.
- All have four rows of metadata we do not need so we can skip them.
- Education and household have no column names so we can add those.
- Sex_age also has no column names but has four columns.
- Education has 244 rows of data, as expected, and then extra data at the end. We can set the maximum number of rows to read at 244.
- Education has 305 and sex_age has 732.
education_raw <- readxl::read_xlsx("./data/education.xlsx",
skip = 4,
col_names = c("Municipality", "Indicator", "Value"),
n_max = 244)
households_raw <- readxl::read_xlsx("./data/households.xlsx",
skip = 4,
col_names = c("Municipality", "Indicator", "Value"),
n_max = 305)
sex_age_raw <- readxl::read_xlsx("./data/sex_age.xlsx",
skip = 4,
col_names = c("Municipality", "Sex", "Age", "Value"),
n_max = 732)It’s generally best to avoid hard-coding limits like this however it simplifies the cleaning and it allows you to confirm the code is loading the expected number of rows.
- In this case the data structure should be robust.
- If there is a change to the data structure, e.g., they added new indicators,you would know that when exporting and the hard coding is easy to find and clear.
Let’s look at the data one by one using both glimpse() and view() to see what might need to be cleaned and re-shaped. What do you notice for each?
Museums
Rows: 29
Columns: 11
$ Location <chr> "tirana - national historical museum", "durrës - archaeolog…
$ `Nov 2018` <dbl> 6306, 855, 1880, 2791, 481, 204, 2088, 767, 134, 593, 836, …
$ `Dec 2018` <dbl> 1191, 324, 117, 1572, 202, 252, 852, 370, 117, 517, 632, 71…
$ `Jan 2019` <dbl> 1016, 95, 138, 912, 176, 180, 768, 232, 87, 73, 289, 593, 4…
$ `Feb 2019` <dbl> 1519, 247, 89, 705, 198, 139, 892, 404, 174, 367, 1116, 962…
$ `Mar 2019` <dbl> 3648, 638, 228, 3101, 758, 221, 2096, 516, 343, 896, 1083, …
$ `Apr 2019` <dbl> 5448, 1501, 410, 6005, 1584, 917, 4840, 964, 230, 467, 1420…
$ `May 2019` <dbl> 7142, 3328, 843, 10352, 4072, 1671, 8944, 1351, 260, 579, 2…
$ `Jun 2019` <dbl> 6004, 2747, 292, 7967, 4682, 1164, 6571, 2185, 0, 0, 1614, …
$ `Jul 2019` <dbl> 7837, 2665, 360, 8364, 3448, 1282, 5618, 549, 53, 0, 1646, …
$ `Aug 2019` <dbl> 10940, 2680, 489, 10659, 4708, 1370, 7281, 1192, 127, 0, 24…- Data is not clean as the site names in the
Locationare lower case and combined with the municipality. - It is not tidy as it has dates as columns.
GDP and Population
Rows: 61
Columns: 4
$ gov_entity <chr> "Dimal", "Pogradec", "Prrenjas", "Kurbin", …
$ population <dbl> 28135, 46070, 18768, 34405, 62232, 51191, 3…
$ general_revenue_per_person <dbl> 14856, 15146, 15466, 16134, 16889, 17127, 1…
$ average_revenue_per_person <dbl> 28307, 28307, 28307, 28307, 28307, 28307, 2…- Data looks clean but the average_revenue_per_person is the same for every municipality.
Schools
Rows: 381
Columns: 9
$ Id <dbl> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 1…
$ School <chr> "Starove", "Sinjë", "Roshnik", "Lapardha", "…
$ Regional_Education_Office <chr> "Berat", "Berat", "Berat", "Berat", "Berat",…
$ Municipality <chr> "Berat", "Berat", "Berat", "Berat", "Berat",…
$ Local_Unit <chr> "Velabisht", "Sinjë", "Roshnik", "Otllak", "…
$ City_Village <chr> "Starove", "Sinjë", "Roshnik", "Lapardha", "…
$ Program_Type <chr> "High School", "High School", "High School",…
$ Dependency_Type <chr> "Classic", "Classic", "Classic", "Classic", …
$ Combined_Separate <chr> "Combined with 9-year school", "Combined wit…- Data looks clean and tidy. It is all character data except for the
Id.
Education
Rows: 244
Columns: 3
$ Municipality <chr> "Berat", NA, NA, NA, "Kuçovë", NA, NA, NA, "Poliçan", NA,…
$ Indicator <chr> "Illiteracy rate: persons 10 years and over", "Educationa…
$ Value <dbl> 2.7, 52.1, 33.9, 13.2, 2.9, 49.1, 36.2, 13.9, 4.8, 56.5, …- The data is not clean as there are many
NAs instead of the values due to the excel structure. - The data is not tidy as all the variables are combined into an
Indicatorscolumn.
Households
Rows: 305
Columns: 3
$ Municipality <chr> "Berat", NA, NA, NA, NA, "Kuçovë", NA, NA, NA, NA, "Poliç…
$ Indicator <chr> "Avarage household size", "Households possessing a washin…
$ Value <dbl> 3.8, 96.3, 71.4, 21.4, 36.9, 3.8, 95.5, 74.4, 14.8, 37.4,…- The data is not clean as there are many
NAs instead of the values due to the excel structure. The word “avarage” inIndicatoris a misspelling. Some names are long. - It is not tidy as all the variables are combined into an
Indicatorcolumn.
Sex and Age
Rows: 732
Columns: 4
$ Municipality <chr> "Belsh", NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, "Ber…
$ Sex <chr> "Total", NA, NA, NA, "Male", NA, NA, NA, "Female", NA, NA…
$ Age <chr> "Total", "0-14", "15-64", "65+", "Total", "0-14", "15-64"…
$ Value <dbl> 17123, 2299, 10537, 4287, 8435, 1195, 5177, 2063, 8688, 1…- The data is not clean as there are many
NAs in two columns instead of the values due to the excel structure. - The data is not tidy. There is a
Totalscategory mixed in with theMaleandFemalein theSexandAgecolumns which could lead to double counting if not careful.
2.4.2 Clean and Reshape the Data to be Tidy
To clean this data, we will go step-by-step following the mantra “Build-a-little, test-a-little”.
- Instead of writing a lot of code at once, we will work on one thing at a time and use the natural pipe,
|>, to connect each step to the next. - It’s easy to do as two key features of almost all tidyverse functions that manipulate data frames are
- They have a data frame as their first argument, and,
- They return a data frame.
By default, the pipe passes the object on the left of the |> to the first argument position of the function on the right (or on the next line) so connecting each step is a series of do this, then do that steps.
At each step you should forecast what you think the resulting output should look like.
Then check what it actually looks like.
- Is it the right number of rows?
- Are the variables in the right places, have the correct class, and are they named correctly?
- Are the values reasonable?
These checks can help you find and debug any errors that occur right away instead of after finding an error many lines later.
Museums
- Let’s start with converting the
Locationnames to Title Case. - We can “mutate” the data frame using
mutatefrom {dplyr} to change a variable value or add a new variable. - Since
Locationhas class character, we will use thestr_to_title()function from {stringr}.
# A tibble: 29 × 11
Location `Nov 2018` `Dec 2018` `Jan 2019` `Feb 2019` `Mar 2019` `Apr 2019`
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Tirana - N… 6306 1191 1016 1519 3648 5448
2 Durrës - A… 855 324 95 247 638 1501
3 Vlorë - Mu… 1880 117 138 89 228 410
4 Krujë - Gj… 2791 1572 912 705 3101 6005
5 Krujë - Et… 481 202 176 198 758 1584
6 Berat - Et… 204 252 180 139 221 917
7 Berat - On… 2088 852 768 892 2096 4840
8 Korçë - Mu… 767 370 232 404 516 964
9 Korçë - Ar… 134 117 87 174 343 230
10 Korçë - Ed… 593 517 73 367 896 467
# ℹ 19 more rows
# ℹ 4 more variables: `May 2019` <dbl>, `Jun 2019` <dbl>, `Jul 2019` <dbl>,
# `Aug 2019` <dbl>Note that we did not save the result but we are checking it interactively. Once we have completed all the steps we can then save it.
- This avoids creating a lot of intermediate variables that will clutter up our environment.
Now let’s separate the the municipality from the site name.
- We can see there is a
-that is in between the two parts we want to separate. - We can use the
separate_wider_delim()from {dplyr}. - Let’s check the help to get the syntax correct.
museums_raw |>
mutate(Location = str_to_title(Location)) |>
separate_wider_delim(Location, " - ", names = c("Municipality", "Site"))# A tibble: 29 × 12
Municipality Site `Nov 2018` `Dec 2018` `Jan 2019` `Feb 2019` `Mar 2019`
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Tirana National… 6306 1191 1016 1519 3648
2 Durrës Archaeol… 855 324 95 247 638
3 Vlorë Museum O… 1880 117 138 89 228
4 Krujë Gjergj K… 2791 1572 912 705 3101
5 Krujë Ethnogra… 481 202 176 198 758
6 Berat Ethnogra… 204 252 180 139 221
7 Berat Onufri I… 2088 852 768 892 2096
8 Korçë Museum O… 767 370 232 404 516
9 Korçë Archaeol… 134 117 87 174 343
10 Korçë Educatio… 593 517 73 367 896
# ℹ 19 more rows
# ℹ 5 more variables: `Apr 2019` <dbl>, `May 2019` <dbl>, `Jun 2019` <dbl>,
# `Jul 2019` <dbl>, `Aug 2019` <dbl>Now let’s fix the columns.
- As you look at the data frame you see it is wider than it should be. We want it to be longer with one column for the dates and one for the values of the number of visitors.
- We can use the “pivot_longer()” function from {tidyr}.
- Let’s look at the help to get the syntax right.
- The tricky part is defining the columns we want to pivot in a way that does not “hard code” the number of columns in case we get more data.
- We could do this in several ways but we notice that all the columns we want to pivot are numeric and they are the only numeric columns.
- This means we can use one of the
tidy-selecthelper functions from {tidyr} that were designed just for this kind of task. They work in many places where we want to select just a subset of columns that have similar characteristics.
- We will repeat our first two steps and pipe into this step.
museums_raw |>
mutate(Location = str_to_title(Location)) |>
separate_wider_delim(Location, " - ", names = c("Municipality", "Site")) |>
pivot_longer(cols = where(is.numeric), names_to = "Date", values_to = "Visitors")# A tibble: 290 × 4
Municipality Site Date Visitors
<chr> <chr> <chr> <dbl>
1 Tirana National Historical Museum Nov 2018 6306
2 Tirana National Historical Museum Dec 2018 1191
3 Tirana National Historical Museum Jan 2019 1016
4 Tirana National Historical Museum Feb 2019 1519
5 Tirana National Historical Museum Mar 2019 3648
6 Tirana National Historical Museum Apr 2019 5448
7 Tirana National Historical Museum May 2019 7142
8 Tirana National Historical Museum Jun 2019 6004
9 Tirana National Historical Museum Jul 2019 7837
10 Tirana National Historical Museum Aug 2019 10940
# ℹ 280 more rowsThis is looking much better. We notice one more fix to make. The Date column is of class character and we want it to be an actual date.
- Character dates don’t sort well and you cannot do math on them.
- Let’s use the
my()the helper functions from {lubridate} to easily convert this.
museums_raw |>
mutate(Location = str_to_title(Location)) |>
separate_wider_delim(Location, " - ", names = c("Municipality", "Site")) |>
pivot_longer(cols = where(is.numeric), names_to = "Date", values_to = "Visitors") |>
mutate(Date = my(Date))# A tibble: 290 × 4
Municipality Site Date Visitors
<chr> <chr> <date> <dbl>
1 Tirana National Historical Museum 2018-11-01 6306
2 Tirana National Historical Museum 2018-12-01 1191
3 Tirana National Historical Museum 2019-01-01 1016
4 Tirana National Historical Museum 2019-02-01 1519
5 Tirana National Historical Museum 2019-03-01 3648
6 Tirana National Historical Museum 2019-04-01 5448
7 Tirana National Historical Museum 2019-05-01 7142
8 Tirana National Historical Museum 2019-06-01 6004
9 Tirana National Historical Museum 2019-07-01 7837
10 Tirana National Historical Museum 2019-08-01 10940
# ℹ 280 more rows- Note: once you get more practice you can do the conversion of the
Datevariable as part of the pivot using thenames_transformargument. This would simply the code for maintenance as it keeps the pivot actions inside the pivot function.
Now that everything looks good, let’s save the data.
- We will give it a new name, but we could just overwrite the old name as well.
We can also save to our data folder for reuse.
We just went through a step-by-step process of cleaning and reshaping the data into a tidy data frame.
- It took a combination of tools from different tidyverse packages.
- We now have a tidy data set we can use for analysis.
The GDP and Population data looks clean and tidy. However, let’s get rid of the last column since it appears to be an error that all the values are the same.
- There are many ways in Base R to remove a column from a data frame. Here are a few to remove the last column in
gdp_pop_raw.
# A tibble: 61 × 3
gov_entity population general_revenue_per_person
<chr> <dbl> <dbl>
1 Dimal 28135 14856
2 Pogradec 46070 15146
3 Prrenjas 18768 15466
4 Kurbin 34405 16134
5 Berat 62232 16889
6 Krujë 51191 17127
7 Kuçovë 31077 17132
8 Belsh 17123 17183
9 Cërrik 25163 17277
10 Devoll 25897 18446
# ℹ 51 more rows# A tibble: 61 × 3
gov_entity population general_revenue_per_person
<chr> <dbl> <dbl>
1 Dimal 28135 14856
2 Pogradec 46070 15146
3 Prrenjas 18768 15466
4 Kurbin 34405 16134
5 Berat 62232 16889
6 Krujë 51191 17127
7 Kuçovë 31077 17132
8 Belsh 17123 17183
9 Cërrik 25163 17277
10 Devoll 25897 18446
# ℹ 51 more rows# A tibble: 61 × 3
gov_entity population general_revenue_per_person
<chr> <dbl> <dbl>
1 Dimal 28135 14856
2 Pogradec 46070 15146
3 Prrenjas 18768 15466
4 Kurbin 34405 16134
5 Berat 62232 16889
6 Krujë 51191 17127
7 Kuçovë 31077 17132
8 Belsh 17123 17183
9 Cërrik 25163 17277
10 Devoll 25897 18446
# ℹ 51 more rows# A tibble: 61 × 3
gov_entity population general_revenue_per_person
<chr> <dbl> <dbl>
1 Dimal 28135 14856
2 Pogradec 46070 15146
3 Prrenjas 18768 15466
4 Kurbin 34405 16134
5 Berat 62232 16889
6 Krujë 51191 17127
7 Kuçovë 31077 17132
8 Belsh 17123 17183
9 Cërrik 25163 17277
10 Devoll 25897 18446
# ℹ 51 more rowsThe select() function in {dplyr} allows you to subset multiple columns from a data frame.
- The
tidyselecthelpers work well withselect().
# A tibble: 61 × 3
gov_entity population general_revenue_per_person
<chr> <dbl> <dbl>
1 Dimal 28135 14856
2 Pogradec 46070 15146
3 Prrenjas 18768 15466
4 Kurbin 34405 16134
5 Berat 62232 16889
6 Krujë 51191 17127
7 Kuçovë 31077 17132
8 Belsh 17123 17183
9 Cërrik 25163 17277
10 Devoll 25897 18446
# ℹ 51 more rowsSchools
- The
schools_rawdata looks good. Let’s just assign a new name to it.
Education
Let’s fix the missing values first so it looks more like a data frame.
- The {tidyr} function
fill()is designed for this situation.
# A tibble: 244 × 3
Municipality Indicator Value
<chr> <chr> <dbl>
1 Berat Illiteracy rate: persons 10 years and over 2.7
2 Berat Educational attainment: primary and lower secondary 52.1
3 Berat Educational attainment: upper secondary 33.9
4 Berat Educational attainment: university 13.2
5 Kuçovë Illiteracy rate: persons 10 years and over 2.9
6 Kuçovë Educational attainment: primary and lower secondary 49.1
7 Kuçovë Educational attainment: upper secondary 36.2
8 Kuçovë Educational attainment: university 13.9
9 Poliçan Illiteracy rate: persons 10 years and over 4.8
10 Poliçan Educational attainment: primary and lower secondary 56.5
# ℹ 234 more rowsNow that we have confirmed it works, we can overwrite the data frame with the new data.
Now let’s fix the variables in the Indicator column.
- Our data frame is too long and we want it wider.
- We can use
pivot_wider()for {tidyr}. - Let’s pipe to
glimpse()to check the results.
Rows: 61
Columns: 5
$ Municipality <chr> "Berat", "Kuçovë…
$ `Illiteracy rate: persons 10 years and over` <dbl> 2.7, 2.9, 4.8, 2…
$ `Educational attainment: primary and lower secondary` <dbl> 52.1, 49.1, 56.5…
$ `Educational attainment: upper secondary` <dbl> 33.9, 36.2, 31.9…
$ `Educational attainment: university` <dbl> 13.2, 13.9, 10.8…We can see the names are quite long and non-syntactic as they have spaces in them.
Let’s save the data and then use the names() function to rename the variables.
education_raw |>
pivot_wider(names_from = Indicator, values_from = Value) ->
education
names(education)[1] "Municipality"
[2] "Illiteracy rate: persons 10 years and over"
[3] "Educational attainment: primary and lower secondary"
[4] "Educational attainment: upper secondary"
[5] "Educational attainment: university" new_names <- c("Municipality", "Illiteracy_rate", "primary_lower_secondary",
"upper_secondary", "university")
names(education) <- new_names
glimpse(education)Rows: 61
Columns: 5
$ Municipality <chr> "Berat", "Kuçovë", "Poliçan", "Skrapar", "Dima…
$ Illiteracy_rate <dbl> 2.7, 2.9, 4.8, 2.4, 2.5, 2.0, 2.7, 5.2, 3.0, 1…
$ primary_lower_secondary <dbl> 52.1, 49.1, 56.5, 52.8, 55.4, 56.4, 56.8, 59.7…
$ upper_secondary <dbl> 33.9, 36.2, 31.9, 33.4, 31.4, 31.1, 29.1, 28.7…
$ university <dbl> 13.2, 13.9, 10.8, 13.2, 12.1, 11.8, 13.6, 10.7…Let’s save the data as a CSV file.
Households
First we can fix the missing data as before.
- Now let’s fix the spelling of “Avarage”.
- We are going to use the
string_replace()function from {stringr}.- We will match (find) the misspelling pattern “Ava” using what is known as a Regular Expression or REGEX or regex.
- The entire second page of the {stringr} cheat sheet is devoted to REGEX as used in R.
- REGEX is a language of its own that is used by many different computer languages to define simple and complex patterns in character strings. As an example, one can use regex to validate a string of characters meets the international standard for URLs.
- Many {stringr} functions have a
patternargument which accepts regex. - For simple regex, you you can use the characters you want to match.
- For more complex patterns, use wildcards or quantifiers or other regex elements to get increasingly complex matches.
# A tibble: 305 × 3
Municipality Indicator Value
<chr> <chr> <dbl>
1 Berat Average household size 3.8
2 Berat Households possessing a washing machine 96.3
3 Berat Households with internet access 71.4
4 Berat Households possessing a computer 21.4
5 Berat Households owning at least one car 36.9
6 Kuçovë Average household size 3.8
7 Kuçovë Households possessing a washing machine 95.5
8 Kuçovë Households with internet access 74.4
9 Kuçovë Households possessing a computer 14.8
10 Kuçovë Households owning at least one car 37.4
# ℹ 295 more rowsNow we can do what we did before and fix the variables being in the Indicator column.
Rows: 61
Columns: 6
$ Municipality <chr> "Berat", "Kuçovë", "Poliçan"…
$ `Avarage household size` <dbl> 3.8, 3.8, 3.7, 3.7, 4.2, 4.6…
$ `Households possessing a washing machine` <dbl> 96.3, 95.5, 95.5, 94.5, 94.5…
$ `Households with internet access` <dbl> 71.4, 74.4, 52.8, 27.7, 80.0…
$ `Households possessing a computer` <dbl> 21.4, 14.8, 10.5, 12.3, 11.7…
$ `Households owning at least one car` <dbl> 36.9, 37.4, 26.3, 25.9, 46.2…Looks good. Let’s save and fix the names.
Show code
[1] "Municipality"
[2] "Avarage household size"
[3] "Households possessing a washing machine"
[4] "Households with internet access"
[5] "Households possessing a computer"
[6] "Households owning at least one car" Show code
Rows: 61
Columns: 6
$ Municipality <chr> "Berat", "Kuçovë", "Poliçan", "Skrapar", "Dimal", …
$ Average_size <dbl> 3.8, 3.8, 3.7, 3.7, 4.2, 4.6, 4.9, 3.7, 3.1, 3.1, …
$ has_washing_machine <dbl> 96.3, 95.5, 95.5, 94.5, 94.5, 92.6, 95.7, 87.7, 90…
$ has_internet <dbl> 71.4, 74.4, 52.8, 27.7, 80.0, 81.3, 85.1, 65.0, 67…
$ has_computer <dbl> 21.4, 14.8, 10.5, 12.3, 11.7, 14.0, 17.4, 10.6, 18…
$ has_car <dbl> 36.9, 37.4, 26.3, 25.9, 46.2, 36.5, 39.2, 33.2, 30…Let’s save the data as a CSV file.
Sex and Age
Let’s fix the missing values first so it looks more like a data frame.
- Let’s check the help to see how we fix two columns at once.
# A tibble: 732 × 4
Municipality Sex Age Value
<chr> <chr> <chr> <dbl>
1 Belsh Total Total 17123
2 Belsh Total 0-14 2299
3 Belsh Total 15-64 10537
4 Belsh Total 65+ 4287
5 Belsh Male Total 8435
6 Belsh Male 0-14 1195
7 Belsh Male 15-64 5177
8 Belsh Male 65+ 2063
9 Belsh Female Total 8688
10 Belsh Female 0-14 1104
# ℹ 722 more rows- Just putting both column names in worked fine.
Now let’s figure out what to do about the totals.
- Let’s first check if the Male and Female numbers add up to the Total for each municipality.
- This is hard to do the way the data is shaped right now.
- Let use a “trick” of pivoting the data wider to separate out the Total, Male and Female into their own columns.
- It is much easier to do math with columns than we rows.
- Now we can create a new column using
mutate()to sum Male and Female and compare.
sex_age_raw |>
fill(Municipality, Sex) |>
pivot_wider(names_from = Sex, values_from = Value) |>
mutate(GT = Male + Female,
is_equal = GT == Total, .after = Total) #|> view()# A tibble: 244 × 7
Municipality Age Total GT is_equal Male Female
<chr> <chr> <dbl> <dbl> <lgl> <dbl> <dbl>
1 Belsh Total 17123 17123 TRUE 8435 8688
2 Belsh 0-14 2299 2299 TRUE 1195 1104
3 Belsh 15-64 10537 10537 TRUE 5177 5360
4 Belsh 65+ 4287 4287 TRUE 2063 2224
5 Berat Total 62232 62232 TRUE 31051 31181
6 Berat 0-14 8799 8799 TRUE 4561 4238
7 Berat 15-64 39011 39011 TRUE 19484 19527
8 Berat 65+ 14422 14422 TRUE 7006 7416
9 Bulqizë Total 26826 26826 TRUE 13608 13218
10 Bulqizë 0-14 4732 4732 TRUE 2477 2255
# ℹ 234 more rowsWe can quickly see that the numbers add up.
We want to keep the Totals now but we want to put the Male and Female back into a Sex column.
sex_age_raw |>
fill(Municipality, Sex) |>
pivot_wider(names_from = Sex, values_from = Value) |>
pivot_longer(cols = c(Male, Female), names_to = "Sex", values_to = "Count")# A tibble: 488 × 5
Municipality Age Total Sex Count
<chr> <chr> <dbl> <chr> <dbl>
1 Belsh Total 17123 Male 8435
2 Belsh Total 17123 Female 8688
3 Belsh 0-14 2299 Male 1195
4 Belsh 0-14 2299 Female 1104
5 Belsh 15-64 10537 Male 5177
6 Belsh 15-64 10537 Female 5360
7 Belsh 65+ 4287 Male 2063
8 Belsh 65+ 4287 Female 2224
9 Berat Total 62232 Male 31051
10 Berat Total 62232 Female 31181
# ℹ 478 more rowsNow let’s repeat with the Age column but now it gets much trickier.
- Let’s stop for a minute and consider how we want to data to look at the end.
- We want columns: Municipality, Sex, Age and then counts for the combination of Sex and Age, and then the counts by Sex, and then the counts by Age, and then the total population for the municipality for all Sexes and Ages. This will take some work.
- It will also create some duplicate data so when we got to use this data, we will want to consider using the
distinct()function from {dplyr} to avoid duplicate counting. - This is where looking at the data will be critical.
- Let’s reuse the data from the previous chunk to get the totals out of the
Sexcolumn. - Now when we try to pivot wider again on
Age, we will (temporarily) have twoTotals column so we usename_repair= minimalto allow that.- Notice this generates a lot of
NAvalues which is fine.
- Notice this generates a lot of
- Now to pivot the
Agecategories back, we will use the tidy-select helpercontainsto get the columns with a-and with a+.- We will also use the
values_drop_na = TRUEto get rid of all theNAs.
- We will also use the
- To reduce confusion we will rename the Totals column to
age_countand relocate it where we want. - Now we will use the
group_by()function from {dplyr} to generate the counts.- This essentially turns the large data frame into a set of smaller data frames for each municipality and sex so we can just sum of the totals.
- Notice the use of the
.after =argument to locate it where we want.
- Now use
group_by()to regroup just by municipality to get the grand total for each municipality. - Now we
ungroup()and it is back to one large data frame with all of our data.
sex_age_raw |>
fill(Municipality, Sex) |>
pivot_wider(names_from = Sex, values_from = Value) |>
pivot_longer(cols = c(Male, Female), names_to = "Sex", values_to = "Count") |>
pivot_wider(names_from = Age, values_from = Count, names_repair = "minimal") |>
pivot_longer(cols = contains(c("-", "+")), names_to = "Age",
values_to = "sex_age_count", values_drop_na = TRUE) |>
rename(age_count = Total) |>
relocate(age_count, .after = sex_age_count) |>
group_by(Municipality, Sex) |>
mutate(sex_count = sum(sex_age_count), .after = sex_age_count) |>
group_by(Municipality) |>
mutate(muni_pop = sum(sex_age_count), .after = age_count) |>
ungroup()# A tibble: 366 × 7
Municipality Sex Age sex_age_count sex_count age_count muni_pop
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 Belsh Male 0-14 1195 8435 2299 17123
2 Belsh Female 0-14 1104 8688 2299 17123
3 Belsh Male 15-64 5177 8435 10537 17123
4 Belsh Female 15-64 5360 8688 10537 17123
5 Belsh Male 65+ 2063 8435 4287 17123
6 Belsh Female 65+ 2224 8688 4287 17123
7 Berat Male 0-14 4561 31051 8799 62232
8 Berat Female 0-14 4238 31181 8799 62232
9 Berat Male 15-64 19484 31051 39011 62232
10 Berat Female 15-64 19527 31181 39011 62232
# ℹ 356 more rowsWe have 6 rows or each of 61 municipalities so the numbers work out.
Now that we are satisfied, let’s save it to a new name that includes “pop” since we created a new variable with the total population for each municipality.
sex_age_raw |>
fill(Municipality, Sex) |>
pivot_wider(names_from = Sex, values_from = Value) |>
pivot_longer(cols = c(Male, Female), names_to = "Sex", values_to = "Count") |>
pivot_wider(names_from = Age, values_from = Count, names_repair = "minimal") |>
pivot_longer(cols = contains(c("-", "+")), names_to = "Age",
values_to = "sex_age_count", values_drop_na = TRUE) |>
rename(age_count = Total) |>
relocate(age_count, .after = sex_age_count) |>
group_by(Municipality, Sex) |>
mutate(sex_count = sum(sex_age_count), .after = sex_age_count) |>
group_by(Municipality) |>
mutate(muni_pop = sum(sex_age_count), .after = age_count) |>
ungroup() ->
sex_age_popLet’s save the data as a CSV file.
All the data has been cleaned and tidied.
We used function from multiple packages and multiple arguments from the functions to minimize the amount of extra code to clean things up.
- This data was fairly clean to begin with.
- The reshaping to adjust where variables are is fairly common.
- The wrangling of the Sex and Age data is also common when there are multiple layers of aggregation in the data.
The {janitor} package can be useful for cleaning variable names and other tasks with really messy data.
2.5 Working with Data Frames
It is common to have large data frames with lots of data in them, but not want to use it all for every operation.
Thus {dplyr} has functions to allow you to manipulate the data even more.
2.5.1 Filter Rows and Select Columns
It’s common to work with a subset of the rows in a data frame.
- While you can do that in Base R with the
[]operator, thefilter()function works as well.
Say we want to filter the culture sites in museums to just those in Vlorë.
- We will use the
==operator for a logical comparison.
# A tibble: 40 × 4
Municipality Site Date Visitors
<chr> <chr> <date> <dbl>
1 Vlorë Museum Of Independence 2018-11-01 1880
2 Vlorë Museum Of Independence 2018-12-01 117
3 Vlorë Museum Of Independence 2019-01-01 138
4 Vlorë Museum Of Independence 2019-02-01 89
5 Vlorë Museum Of Independence 2019-03-01 228
6 Vlorë Museum Of Independence 2019-04-01 410
7 Vlorë Museum Of Independence 2019-05-01 843
8 Vlorë Museum Of Independence 2019-06-01 292
9 Vlorë Museum Of Independence 2019-07-01 360
10 Vlorë Museum Of Independence 2019-08-01 489
# ℹ 30 more rowsNow let’s say we only want for the November and December 2018, we can combine the logical tests.
- The
,serves as the logicalAND.
# A tibble: 8 × 4
Municipality Site Date Visitors
<chr> <chr> <date> <dbl>
1 Vlorë Museum Of Independence 2018-11-01 1880
2 Vlorë Museum Of Independence 2018-12-01 117
3 Vlorë Castle Of Kaninë 2018-11-01 20
4 Vlorë Castle Of Kaninë 2018-12-01 20
5 Vlorë Amantia Archaeological Park 2018-11-01 0
6 Vlorë Amantia Archaeological Park 2018-12-01 70
7 Vlorë Orikum Archaeological Park 2018-11-01 46
8 Vlorë Orikum Archaeological Park 2018-12-01 27What if we wanted the sites in Vlorë and Tirana.
- Notice the use of
%in%instead of==since we have more than one item to compare.
# A tibble: 12 × 4
Municipality Site Date Visitors
<chr> <chr> <date> <dbl>
1 Tirana National Historical Museum 2018-11-01 6306
2 Tirana National Historical Museum 2018-12-01 1191
3 Vlorë Museum Of Independence 2018-11-01 1880
4 Vlorë Museum Of Independence 2018-12-01 117
5 Tirana House Of Leaves (Surveillance Museum) 2018-11-01 1336
6 Tirana House Of Leaves (Surveillance Museum) 2018-12-01 717
7 Vlorë Castle Of Kaninë 2018-11-01 20
8 Vlorë Castle Of Kaninë 2018-12-01 20
9 Vlorë Amantia Archaeological Park 2018-11-01 0
10 Vlorë Amantia Archaeological Park 2018-12-01 70
11 Vlorë Orikum Archaeological Park 2018-11-01 46
12 Vlorë Orikum Archaeological Park 2018-12-01 27What if we wanted a list of just the sites for the two cities?
# A tibble: 60 × 4
Municipality Site Date Visitors
<chr> <chr> <date> <dbl>
1 Tirana National Historical Museum 2018-11-01 6306
2 Tirana National Historical Museum 2018-12-01 1191
3 Tirana National Historical Museum 2019-01-01 1016
4 Tirana National Historical Museum 2019-02-01 1519
5 Tirana National Historical Museum 2019-03-01 3648
6 Tirana National Historical Museum 2019-04-01 5448
7 Tirana National Historical Museum 2019-05-01 7142
8 Tirana National Historical Museum 2019-06-01 6004
9 Tirana National Historical Museum 2019-07-01 7837
10 Tirana National Historical Museum 2019-08-01 10940
# ℹ 50 more rowsThat did not give us what we want as every row is distinct. This is where select() comes to play.
- The
select()function from {dplyr} is for choosing (subsetting) the columns you want (or don’t want) in the result.- In tidyverse functions we can usually use the variable names.
- We have already seen examples using the tidy-select functions for more complex selections.
- Let’s select the just the first two columns before filtering.
- The
distinct()ensures we don’t get duplicates.
- The
museums |>
select(Municipality, Site) |>
filter(Municipality %in% c("Vlorë", "Tirana")) |>
distinct()# A tibble: 6 × 2
Municipality Site
<chr> <chr>
1 Tirana National Historical Museum
2 Vlorë Museum Of Independence
3 Tirana House Of Leaves (Surveillance Museum)
4 Vlorë Castle Of Kaninë
5 Vlorë Amantia Archaeological Park
6 Vlorë Orikum Archaeological Park What if we want the sites and months with the top 5 or bottom five visitors?
- This is where the slice functions come to play.
# A tibble: 10 × 4
Municipality Site Date Visitors
<chr> <chr> <date> <dbl>
1 Sarandë Butrint National Park 2019-07-01 46506
2 Sarandë Butrint National Park 2019-08-01 46506
3 Gjirokastër Castle Of Gjirokastër 2019-08-01 30191
4 Sarandë Butrint National Park 2019-06-01 28087
5 Sarandë Butrint National Park 2019-05-01 28000
6 Berat Castle Of Berat 2019-08-01 25686
7 Fier Apollonia Archaeological Park 2019-05-01 19088
8 Berat Castle Of Berat 2019-05-01 18305
9 Shkodër Shkodër Archaeological Park 2019-07-01 16548
10 Shkodër Shkodër Archaeological Park 2019-08-01 16548# A tibble: 15 × 4
Municipality Site Date Visitors
<chr> <chr> <date> <dbl>
1 Korçë Archaeological Museum 2019-06-01 0
2 Korçë Education Museum 2019-06-01 0
3 Korçë Education Museum 2019-07-01 0
4 Korçë Education Museum 2019-08-01 0
5 Sarandë Monastery Of 40 Saints 2018-12-01 0
6 Sarandë Monastery Of 40 Saints 2019-01-01 0
7 Finiq Monastery Of Mesopotam 2018-11-01 0
8 Finiq Monastery Of Mesopotam 2018-12-01 0
9 Finiq Monastery Of Mesopotam 2019-01-01 0
10 Finiq Monastery Of Mesopotam 2019-02-01 0
11 Vlorë Amantia Archaeological Park 2018-11-01 0
12 Finiq Finiq Archaeological Park 2018-11-01 0
13 Gjirokastër Antigone Archaeological Park 2018-12-01 0
14 Gjirokastër Antigone Archaeological Park 2019-01-01 0
15 Gjirokastër Antigone Archaeological Park 2019-02-01 0If we want to sort the data, (temporarily) we can use arrange().
- This does not actually change how the data is sorted in the saved data frame unless it is saved.
# A tibble: 290 × 4
Municipality Site Date Visitors
<chr> <chr> <date> <dbl>
1 Vlorë Amantia Archaeological Park 2018-11-01 0
2 Vlorë Amantia Archaeological Park 2018-12-01 70
3 Vlorë Amantia Archaeological Park 2019-01-01 5
4 Vlorë Amantia Archaeological Park 2019-02-01 23
5 Vlorë Amantia Archaeological Park 2019-03-01 38
6 Vlorë Amantia Archaeological Park 2019-04-01 135
7 Vlorë Amantia Archaeological Park 2019-05-01 150
8 Vlorë Amantia Archaeological Park 2019-06-01 363
9 Vlorë Amantia Archaeological Park 2019-07-01 70
10 Vlorë Amantia Archaeological Park 2019-08-01 70
# ℹ 280 more rows# A tibble: 290 × 4
Municipality Site Date Visitors
<chr> <chr> <date> <dbl>
1 Vlorë Amantia Archaeological Park 2019-06-01 363
2 Vlorë Amantia Archaeological Park 2019-05-01 150
3 Vlorë Amantia Archaeological Park 2019-04-01 135
4 Vlorë Amantia Archaeological Park 2018-12-01 70
5 Vlorë Amantia Archaeological Park 2019-07-01 70
6 Vlorë Amantia Archaeological Park 2019-08-01 70
7 Vlorë Amantia Archaeological Park 2019-03-01 38
8 Vlorë Amantia Archaeological Park 2019-02-01 23
9 Vlorë Amantia Archaeological Park 2019-01-01 5
10 Vlorë Amantia Archaeological Park 2018-11-01 0
# ℹ 280 more rows2.5.2 Summarizing Data
We have already seen some examples of summarizing data as part of a mutate() function.
- When using
mutate(), every row is returned so there may be duplicate data.
If we want to summarize a data frame where we reduce the number of rows, we use summarize().
Consider the schools data frame. We can create a summarized data frame that shows the number of schools of each type in each municipality.
- First group by
MunicipalityandProgam_Type. - Then we want to create a new variable called
countwhich is the number of schools of each type. - The
.groups= "dropremoves the second layer of grouping assummarize()removes the first by default.
# A tibble: 78 × 3
Municipality Program_Type count
<chr> <chr> <int>
1 Belsh High School 3
2 Berat High School 8
3 Berat Specialized High School 1
4 Bulqizë High School 6
5 Cërrik High School 4
6 Delvinë High School 1
7 Devoll High School 5
8 Dibër High School 8
9 Dibër Short-term 1
10 Divjake High School 7
# ℹ 68 more rows- Now we only have 78 rows down from 381.
Let’s save this in case we want to use it later.
# A tibble: 61 × 7
Municipality Illiteracy_rate primary_lower_secondary upper_secondary
<chr> <dbl> <dbl> <dbl>
1 Berat 2.7 52.1 33.9
2 Kuçovë 2.9 49.1 36.2
3 Poliçan 4.8 56.5 31.9
4 Skrapar 2.4 52.8 33.4
5 Dimal 2.5 55.4 31.4
6 Bulqizë 2 56.4 31.1
7 Dibër 2.7 56.8 29.1
8 Klos 5.2 59.7 28.7
9 Mat 3 49.7 36.3
10 Durrës 1.7 44 36.3
# ℹ 51 more rows
# ℹ 3 more variables: university <dbl>, population <dbl>,
# general_revenue_per_person <dbl>2.6 Working with Multiple Data Frames
We often find ourselves with data in multiple data frames that we would like in one data frame.
- This is very common in data science, especially when working with data from a relational database that uses lots of tables to minimize the storage of redundant data.
- Many computer languages have some version of the join functions that are present in Structured Query Language (SQL).
- They may be called different kinds of joins or more generic terms such as merge or melt.
The {dplyr} package uses multiple join functions. We will discuss three commonly used functions.
left_join()- a mutating joinsemi_join()- a filtering joinanti-join()- a filtering join
When discussing joins it helps to use pictures as in Figure 2.1
2.6.1 Left Join
The goal of a left join is to add variables from the data frame on the right (Y) to the data frame on the left (X) without duplicating any columns or adjusting the number of rows.
- What makes joins special is the data in the two data frames does not have to be sorted the same.
- A join operates on the basis of “key” fields which allow for matching a Key field in the X data frame to a corresponding “foreign” key field in the Y data frame.
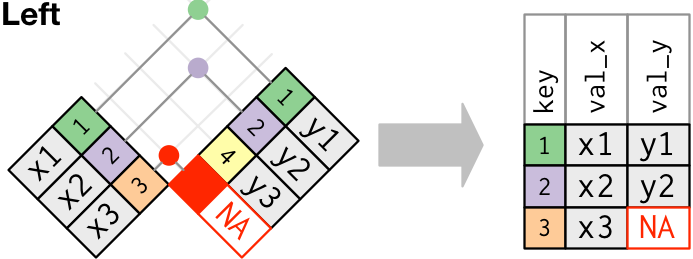
A left join looks like Figure 2.2 where the key fields 1 and 2 match so the values from the right data frame for those fields are added and the value for key field 3 gets an NA.
- Note, no rows are added or removed from the Left data frame.
Let’s add variables from education (right) to the households data frame (left).
Rows: 61
Columns: 10
$ Municipality <chr> "Berat", "Kuçovë", "Poliçan", "Skrapar", "Dima…
$ Average_size <dbl> 3.8, 3.8, 3.7, 3.7, 4.2, 4.6, 4.9, 3.7, 3.1, 3…
$ has_washing_machine <dbl> 96.3, 95.5, 95.5, 94.5, 94.5, 92.6, 95.7, 87.7…
$ has_internet <dbl> 71.4, 74.4, 52.8, 27.7, 80.0, 81.3, 85.1, 65.0…
$ has_computer <dbl> 21.4, 14.8, 10.5, 12.3, 11.7, 14.0, 17.4, 10.6…
$ has_car <dbl> 36.9, 37.4, 26.3, 25.9, 46.2, 36.5, 39.2, 33.2…
$ Illiteracy_rate <dbl> 2.7, 2.9, 4.8, 2.4, 2.5, 2.0, 2.7, 5.2, 3.0, 1…
$ primary_lower_secondary <dbl> 52.1, 49.1, 56.5, 52.8, 55.4, 56.4, 56.8, 59.7…
$ upper_secondary <dbl> 33.9, 36.2, 31.9, 33.4, 31.4, 31.1, 29.1, 28.7…
$ university <dbl> 13.2, 13.9, 10.8, 13.2, 12.1, 11.8, 13.6, 10.7…- We get a message that the function found a common key field
Municipalityin both data frames and used that for the join.
Let’s try to add the variables from gdp_pop to our larger data frame.
Error in `left_join()`:
! `by` must be supplied when `x` and `y` have no common variables.
ℹ Use `cross_join()` to perform a cross-join.- Now we get an error because it cannot find common key fields.
- When we look at the data, we can see that
gdp_pophas municipalities but they have the variable namegov_entity. - We could rename the variable but that might mess up other places where we use that data.
- It is far better to use the
join_by()function we saw in the first message.join_by()allows us to specify how to connect the key fields in the two data frames.
left_join(households, education) |>
left_join(gdp_pop, join_by("Municipality" == "gov_entity")) |>
glimpse()Rows: 61
Columns: 12
$ Municipality <chr> "Berat", "Kuçovë", "Poliçan", "Skrapar", "D…
$ Average_size <dbl> 3.8, 3.8, 3.7, 3.7, 4.2, 4.6, 4.9, 3.7, 3.1…
$ has_washing_machine <dbl> 96.3, 95.5, 95.5, 94.5, 94.5, 92.6, 95.7, 8…
$ has_internet <dbl> 71.4, 74.4, 52.8, 27.7, 80.0, 81.3, 85.1, 6…
$ has_computer <dbl> 21.4, 14.8, 10.5, 12.3, 11.7, 14.0, 17.4, 1…
$ has_car <dbl> 36.9, 37.4, 26.3, 25.9, 46.2, 36.5, 39.2, 3…
$ Illiteracy_rate <dbl> 2.7, 2.9, 4.8, 2.4, 2.5, 2.0, 2.7, 5.2, 3.0…
$ primary_lower_secondary <dbl> 52.1, 49.1, 56.5, 52.8, 55.4, 56.4, 56.8, 5…
$ upper_secondary <dbl> 33.9, 36.2, 31.9, 33.4, 31.4, 31.1, 29.1, 2…
$ university <dbl> 13.2, 13.9, 10.8, 13.2, 12.1, 11.8, 13.6, 1…
$ population <dbl> 62232, 31077, 8762, 10750, 28135, 26826, 50…
$ general_revenue_per_person <dbl> 16889, 17132, 98395, 32924, 14856, 21076, 1…- Now this works fine.
Let’s save this to a new name and to our data folder.
Having all this data in one place makes it easy to do some plots (more in the next chapters).
house_ed_gdp_df |>
ggplot(aes(x = Illiteracy_rate, y = general_revenue_per_person)) +
geom_point() +
labs(title = "Comparing Illiterarcy Rate and General Revenue Person for Municipalities",
caption = "Data from Albania opendata.gv.al and INSTAT for 2023") +
xlab("Illiteracy Rate") +
ylab("General Revenue per Person") +
scale_y_log10() +
geom_smooth(se = FALSE, linetype = 2, color = "red") +
geom_smooth( method = "lm", se = FALSE)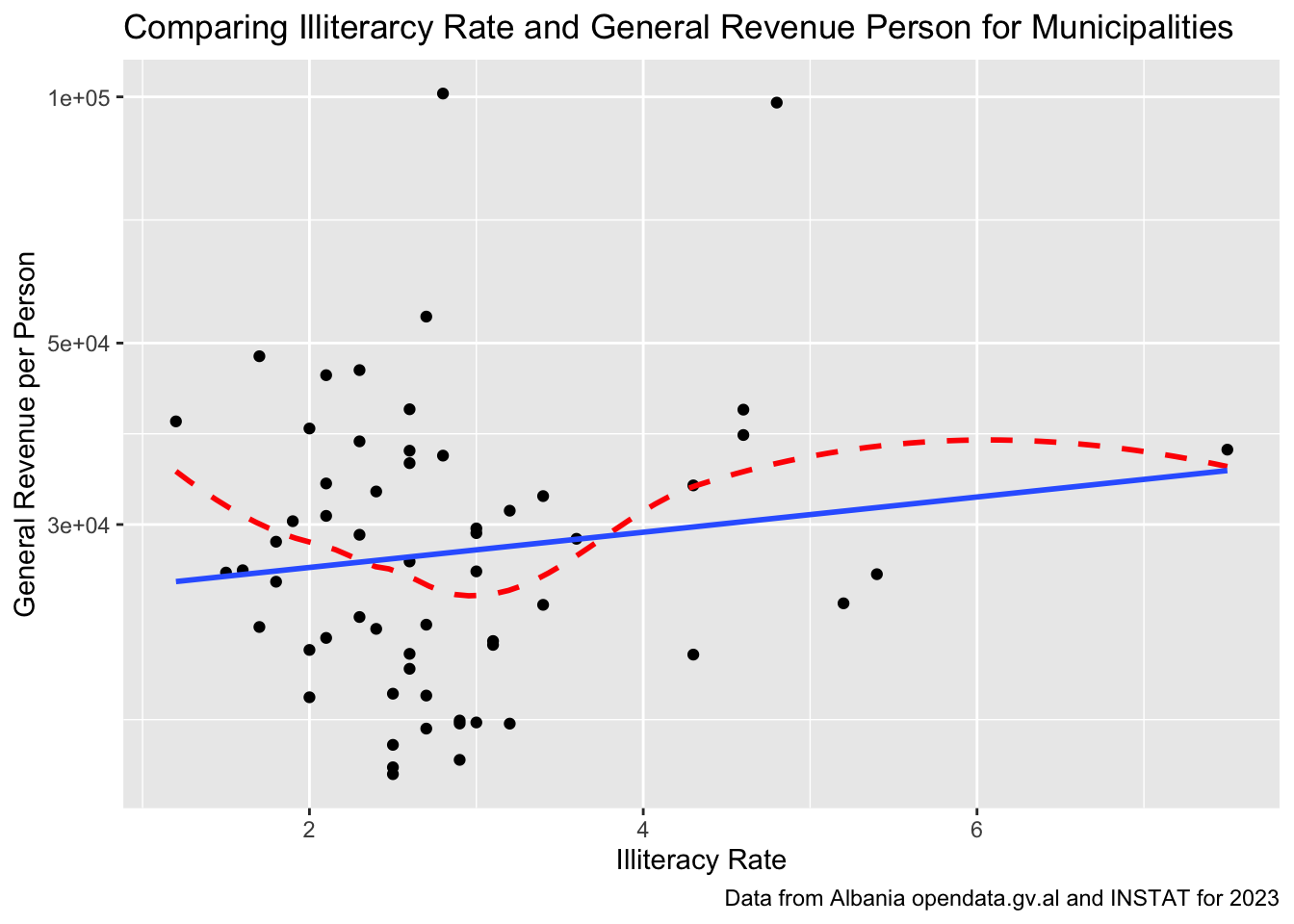
How would you interpret this graph? Do you believe it?
There are other mutating joins such as the Full join, the right join and the inner join.
- A Right join is just a mirror version of the left join so you can usually do a left join.
- Both full joins and inner joins can lead to issues with your data so there is rarely a need for them in data science work.
2.6.2 Filtering Joins
The goals for a filtering join are not about adding variables to a data frame but about removing rows in the left (X) data frame based on the data in the right (Y) data frame.
semi_join(): returns all rows from x with a match in y (deleting those without a match)anti_join(): returns all rows from x without a match in y (deleting those that do match)
The look like
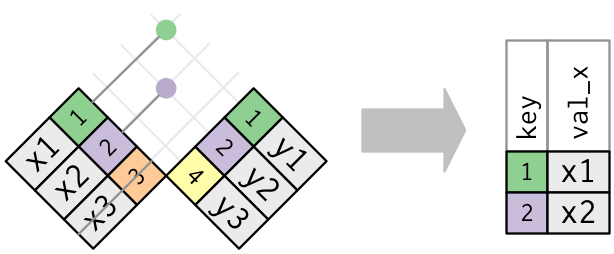
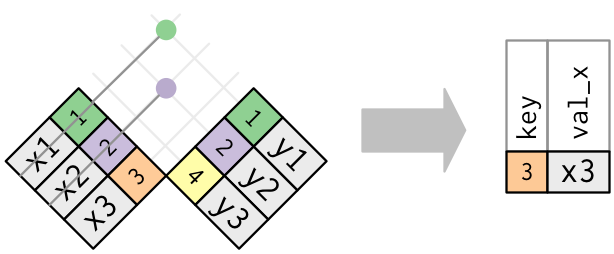
Say we want to find those municipalities that have a cultural site and look at their household data.
# A tibble: 12 × 6
Municipality Average_size has_washing_machine has_internet has_computer
<chr> <dbl> <dbl> <dbl> <dbl>
1 Berat 3.8 96.3 71.4 21.4
2 Durrës 3.1 97.6 77.1 29.2
3 Fier 2.9 95.9 73.5 19.6
4 Finiq 2.7 94.6 32.1 7.8
5 Gjirokastër 2.9 97.3 67.7 23.7
6 Himarë 2.5 95.5 44.5 13.3
7 Korçë 2.8 96.4 80 33.4
8 Krujë 3.7 95 65.3 18.2
9 Lezhë 3.2 95.9 76.2 20.4
10 Sarandë 2.8 97.8 78.3 30.3
11 Shkodër 3.1 95.3 78.1 28.5
12 Vlorë 2.7 97.4 67.8 23.8
# ℹ 1 more variable: has_car <dbl>Only 12 Municipalities have cultural sites.
Does this make sense? There should be 13??
# A tibble: 13 × 1
Municipality
<chr>
1 Tirana
2 Durrës
3 Vlorë
4 Krujë
5 Berat
6 Korçë
7 Shkodër
8 Gjirokastër
9 Himarë
10 Sarandë
11 Finiq
12 Fier
13 Lezhë Now we want to find those that do Not have a cultural site
# A tibble: 49 × 6
Municipality Average_size has_washing_machine has_internet has_computer
<chr> <dbl> <dbl> <dbl> <dbl>
1 Belsh 3.8 94.1 84.6 9.5
2 Bulqizë 4.6 92.6 81.3 14
3 Cërrik 3.9 93.9 80.5 11.2
4 Delvinë 2.8 95 68.7 16.5
5 Devoll 3.9 94.6 79.1 18.4
6 Dibër 4.9 95.7 85.1 17.4
7 Dimal 4.2 94.5 80 11.7
8 Divjakë 3.1 95.7 82 13.6
9 Dropull 2.8 97.4 29.7 13
10 Elbasan 3.1 95.9 77.1 25.1
# ℹ 39 more rows
# ℹ 1 more variable: has_car <dbl>What is going on??
Show code
# A tibble: 13 × 6
Municipality Average_size has_washing_machine has_internet has_computer
<chr> <dbl> <dbl> <dbl> <dbl>
1 Berat 3.8 96.3 71.4 21.4
2 Durrës 3.1 97.6 77.1 29.2
3 Fier 2.9 95.9 73.5 19.6
4 Finiq 2.7 94.6 32.1 7.8
5 Gjirokastër 2.9 97.3 67.7 23.7
6 Himarë 2.5 95.5 44.5 13.3
7 Korçë 2.8 96.4 80 33.4
8 Krujë 3.7 95 65.3 18.2
9 Lezhë 3.2 95.9 76.2 20.4
10 Sarandë 2.8 97.8 78.3 30.3
11 Shkodër 3.1 95.3 78.1 28.5
12 Tiranë 2.9 98 80 49.3
13 Vlorë 2.7 97.4 67.8 23.8
# ℹ 1 more variable: has_car <dbl>Bottom line: Always check your data:)
2.7 Creating a New Variable
We saw earlier that we could create a new summary variable for population from the variables in the data frame and add it to the data frame.
Now let’s create a new variable for one data frame based on data in a second data frame.
Let’s add a variable about whether a municipality has a cultural site and add it to the
house_ed_gdp_dfdata frame.Let’s call the new variable
culture_siteand it will have two values:has_siteandno_site.- This is a categorical variable (only a limited number of discrete possible values) so we will convert it to a factor.
Look at the help for
if_else()andas.factor().
house_ed_gdp_df |>
mutate(culture_site = if_else(
Municipality %in% semi_join(households, museums)$Municipality,
"has_site", "no_site"
), .after = Municipality
) |>
mutate(culture_site = as.factor(culture_site))# A tibble: 61 × 13
Municipality culture_site Average_size has_washing_machine has_internet
<chr> <fct> <dbl> <dbl> <dbl>
1 Berat has_site 3.8 96.3 71.4
2 Kuçovë no_site 3.8 95.5 74.4
3 Poliçan no_site 3.7 95.5 52.8
4 Skrapar no_site 3.7 94.5 27.7
5 Dimal no_site 4.2 94.5 80
6 Bulqizë no_site 4.6 92.6 81.3
7 Dibër no_site 4.9 95.7 85.1
8 Klos no_site 3.7 87.7 65
9 Mat no_site 3.1 90.9 67.5
10 Durrës has_site 3.1 97.6 77.1
# ℹ 51 more rows
# ℹ 8 more variables: has_computer <dbl>, has_car <dbl>, Illiteracy_rate <dbl>,
# primary_lower_secondary <dbl>, upper_secondary <dbl>, university <dbl>,
# population <dbl>, general_revenue_per_person <dbl>This appears to be working so let’s save to the data frame and rewrite to the data folder.
- Note: when stored as a CSV file, it loses it’s class as a factor but that can be recreated upon reading it in.
You can check the levels of a factor with the levels() function.
Factors are commonly used for trying to see if there is a difference between two or more categories.
house_ed_gdp_df |>
ggplot(aes(x = culture_site, y = population)) +
geom_boxplot() +
# scale_y_log10() +
ggtitle("Municipalities with culture sites have a higher median population")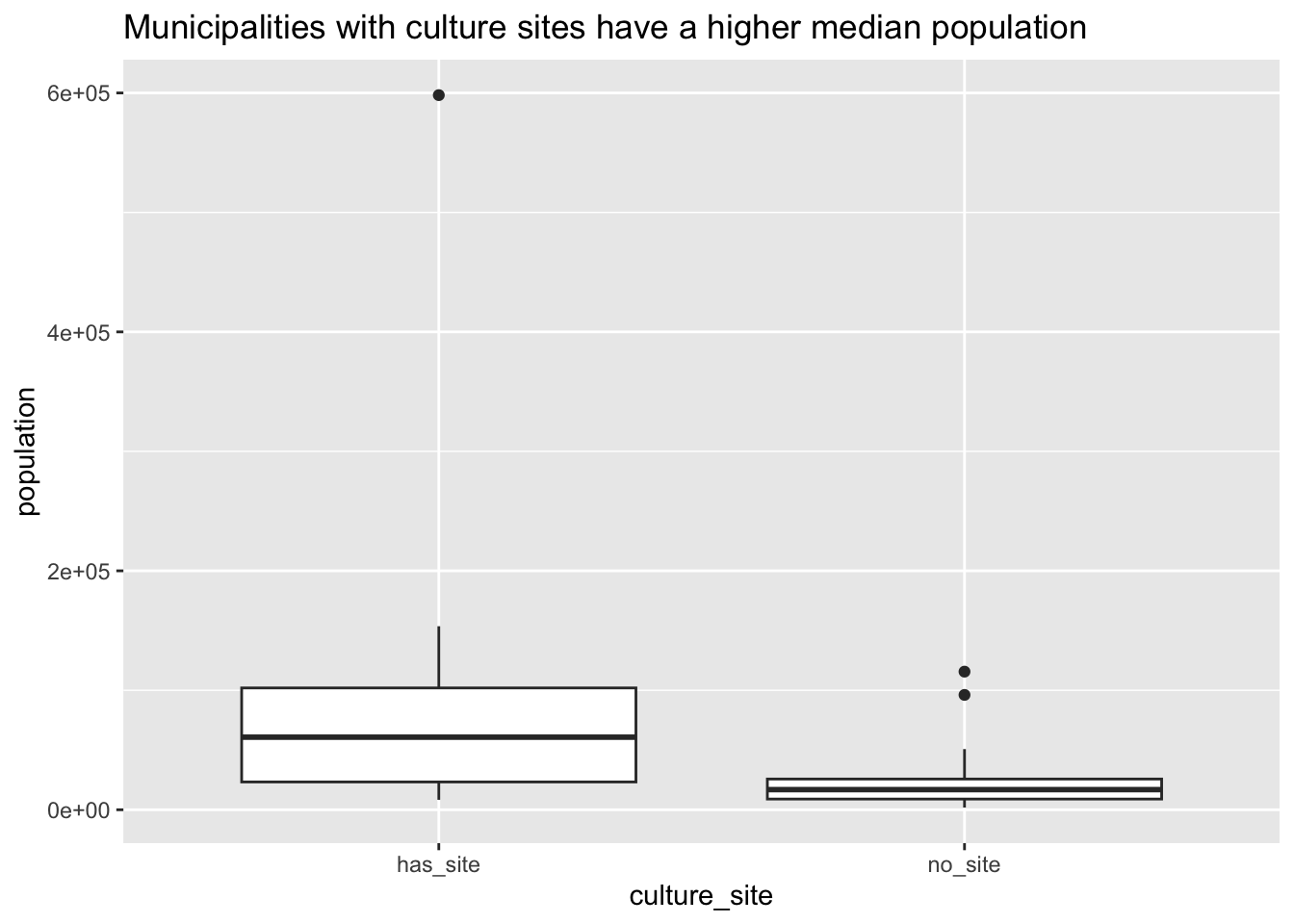
- Here we can see there is a clear difference in the median of the populations between municipalities that have a site and those that do not.
2.8 Summary
You have had a quick exposure to multiple tidyverse functions for manipulating data.
- There are more functions than we had time to cover.
- The goal was to give you some idea of the kinds of problems that can occur and the many ways data scientists can combine functions in creative strategies to solve a data cleaning and shaping problem.
A few keys to success:
- Think about what you want your data to look like.
- Use the Help function to understand how to use the function arguments to solve common problems.
- Use the pipe to help build-a-little, test-a-little in a reproducible and clear way.
- Forecast your results and check your data.
- As always, save your work early and often:).